Please be patient as the page loads
|
NALP7_HUMAN
|
||||||
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NALP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990483 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 1.42253e-146 (rank : 3) | NC score | 0.970727 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 8.69046e-120 (rank : 4) | NC score | 0.966937 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 1.48236e-119 (rank : 5) | NC score | 0.970014 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 2.6227e-108 (rank : 6) | NC score | 0.959536 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 3.42535e-108 (rank : 7) | NC score | 0.960325 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 2.89973e-107 (rank : 8) | NC score | 0.960195 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 2.45477e-106 (rank : 9) | NC score | 0.888263 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 2.37764e-101 (rank : 10) | NC score | 0.950418 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 5.15425e-80 (rank : 11) | NC score | 0.944051 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 3.34088e-79 (rank : 12) | NC score | 0.915250 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 1.84173e-61 (rank : 13) | NC score | 0.934865 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| θ value | 1.19377e-60 (rank : 14) | NC score | 0.933698 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 6.34462e-54 (rank : 15) | NC score | 0.903836 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 1.61718e-49 (rank : 16) | NC score | 0.901681 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
RINI_MOUSE
|
||||||
| θ value | 1.24399e-33 (rank : 17) | NC score | 0.844254 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
RINI_HUMAN
|
||||||
| θ value | 1.92365e-26 (rank : 18) | NC score | 0.843461 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 1.05554e-24 (rank : 19) | NC score | 0.745346 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 1.29031e-22 (rank : 20) | NC score | 0.699467 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 2.20094e-22 (rank : 21) | NC score | 0.708218 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 22) | NC score | 0.628721 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 1.68911e-14 (rank : 23) | NC score | 0.720065 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 3.76295e-14 (rank : 24) | NC score | 0.644568 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 25) | NC score | 0.547993 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 26) | NC score | 0.279722 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 27) | NC score | 0.254121 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 28) | NC score | 0.255954 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 2.88788e-06 (rank : 29) | NC score | 0.248712 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 2.44474e-05 (rank : 30) | NC score | 0.505973 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 31) | NC score | 0.259416 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 32) | NC score | 0.239949 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
LRC31_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 33) | NC score | 0.486732 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
ASC_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 34) | NC score | 0.292250 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 35) | NC score | 0.355264 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.005364 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
NVL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.020852 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PEO1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 38) | NC score | 0.029406 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
CF154_HUMAN
|
||||||
| θ value | 0.813845 (rank : 39) | NC score | 0.469071 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.081250 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC45_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.160165 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
PEX6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.020078 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.020059 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.001893 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
ZNF35_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | -0.003495 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13682, Q96D01 | Gene names | ZNF35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 35 (Zinc finger protein HF.10). | |||||
|
ASC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.224778 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
PEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.026117 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
PNPO_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.015712 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91XF0 | Gene names | Pnpo | |||
|
Domain Architecture |
|
|||||
| Description | Pyridoxine-5'-phosphate oxidase (EC 1.4.3.5) (Pyridoxamine-phosphate oxidase). | |||||
|
TRP13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.025003 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
BMS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.021475 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.029716 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
DHX57_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.006527 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.006496 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
LRIG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | -0.000680 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P1C6, Q6ZPE6, Q8BRF0, Q8BS51, Q8C5E4 | Gene names | Lrig3, Kiaa3016 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 3 precursor. | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.141454 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
TRP13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.023737 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
VPS4B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.011314 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.015981 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.001339 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CLPX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.022344 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
DIRA3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.007173 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |
|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
KGUA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.010065 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64520 | Gene names | Guk1, Gmk | |||
|
Domain Architecture |
|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
NVL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.016579 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PRS10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.010805 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.010805 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
BIRC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.057956 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.058920 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.052415 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052038 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.054412 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |
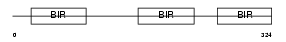
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
BIRC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.055585 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |
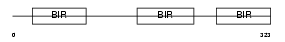
|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.052462 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |
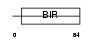
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
BIRC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.051538 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |
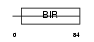
|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.117084 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
CD14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.050947 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |
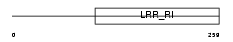
|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
LRC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.082107 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.148034 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MAPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052734 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
RGP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.301098 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.286968 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.990483 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.970727 (rank : 3) | θ value | 1.42253e-146 (rank : 3) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.970014 (rank : 4) | θ value | 1.48236e-119 (rank : 5) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.966937 (rank : 5) | θ value | 8.69046e-120 (rank : 4) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.960325 (rank : 6) | θ value | 3.42535e-108 (rank : 7) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.960195 (rank : 7) | θ value | 2.89973e-107 (rank : 8) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.959536 (rank : 8) | θ value | 2.6227e-108 (rank : 6) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.950418 (rank : 9) | θ value | 2.37764e-101 (rank : 10) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.944051 (rank : 10) | θ value | 5.15425e-80 (rank : 11) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.934865 (rank : 11) | θ value | 1.84173e-61 (rank : 13) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.933698 (rank : 12) | θ value | 1.19377e-60 (rank : 14) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.915250 (rank : 13) | θ value | 3.34088e-79 (rank : 12) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.903836 (rank : 14) | θ value | 6.34462e-54 (rank : 15) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.901681 (rank : 15) | θ value | 1.61718e-49 (rank : 16) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.888263 (rank : 16) | θ value | 2.45477e-106 (rank : 9) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.844254 (rank : 17) | θ value | 1.24399e-33 (rank : 17) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.843461 (rank : 18) | θ value | 1.92365e-26 (rank : 18) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.745346 (rank : 19) | θ value | 1.05554e-24 (rank : 19) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.720065 (rank : 20) | θ value | 1.68911e-14 (rank : 23) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.708218 (rank : 21) | θ value | 2.20094e-22 (rank : 21) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.699467 (rank : 22) | θ value | 1.29031e-22 (rank : 20) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.644568 (rank : 23) | θ value | 3.76295e-14 (rank : 24) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.628721 (rank : 24) | θ value | 1.058e-16 (rank : 22) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.547993 (rank : 25) | θ value | 3.89403e-11 (rank : 25) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.505973 (rank : 26) | θ value | 2.44474e-05 (rank : 30) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.486732 (rank : 27) | θ value | 0.00102713 (rank : 33) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.469071 (rank : 28) | θ value | 0.813845 (rank : 39) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.355264 (rank : 29) | θ value | 0.0193708 (rank : 35) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.301098 (rank : 30) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.292250 (rank : 31) | θ value | 0.00665767 (rank : 34) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.286968 (rank : 32) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.279722 (rank : 33) | θ value | 1.29631e-06 (rank : 26) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.259416 (rank : 34) | θ value | 4.1701e-05 (rank : 31) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.255954 (rank : 35) | θ value | 2.88788e-06 (rank : 28) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.254121 (rank : 36) | θ value | 1.69304e-06 (rank : 27) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.248712 (rank : 37) | θ value | 2.88788e-06 (rank : 29) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.239949 (rank : 38) | θ value | 0.000461057 (rank : 32) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
ASC_HUMAN
|
||||||
| NC score | 0.224778 (rank : 39) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.160165 (rank : 40) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.148034 (rank : 41) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.141454 (rank : 42) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.117084 (rank : 43) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
LRC14_HUMAN
|
||||||
| NC score | 0.082107 (rank : 44) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.081250 (rank : 45) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.058920 (rank : 46) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
BIRC2_HUMAN
|
||||||
| NC score | 0.057956 (rank : 47) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13490, Q16516, Q4TTG0 | Gene names | BIRC2, API1, IAP2, MIHB | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (HIAP2) (HIAP-2) (C-IAP1) (TNFR2-TRAF signaling complex protein 2) (IAP homolog B). | |||||
|
BIRC4_MOUSE
|
||||||
| NC score | 0.055585 (rank : 48) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60989, O08865 | Gene names | Birc4, Aipa, Api3, Miha, Xiap | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP homolog A) (MIAP3) (MIAP-3). | |||||
|
BIRC4_HUMAN
|
||||||
| NC score | 0.054412 (rank : 49) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P98170, Q9NQ14 | Gene names | BIRC4, API3, IAP3, XIAP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 4 (Inhibitor of apoptosis protein 3) (X-linked inhibitor of apoptosis protein) (X-linked IAP) (IAP-like protein) (HILP). | |||||
|
MAPE_HUMAN
|
||||||
| NC score | 0.052734 (rank : 50) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78395, O43481, Q8IXN8 | Gene names | PRAME, MAPE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma antigen preferentially expressed in tumors (Preferentially expressed antigen of melanoma) (OPA-interacting protein 4) (OIP4). | |||||
|
BIRC5_HUMAN
|
||||||
| NC score | 0.052462 (rank : 51) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15392, Q2I3N8, Q4VGX0, Q53F61, Q5MGC6, Q6FHL2, Q75SP2, Q9P2W8 | Gene names | BIRC5, API4, IAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4). | |||||
|
BIRC3_HUMAN
|
||||||
| NC score | 0.052415 (rank : 52) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q13489, Q16628, Q9HC27, Q9UP46 | Gene names | BIRC3, API2, IAP1, MIHC | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (HIAP1) (HIAP-1) (C-IAP2) (TNFR2-TRAF signaling complex protein 1) (IAP homolog C) (Apoptosis inhibitor 2) (API2). | |||||
|
BIRC3_MOUSE
|
||||||
| NC score | 0.052038 (rank : 53) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08863 | Gene names | Birc3, Birc2, Iap1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 3 (Inhibitor of apoptosis protein 1) (MIAP1) (MIAP-1). | |||||
|
BIRC5_MOUSE
|
||||||
| NC score | 0.051538 (rank : 54) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70201, Q923F7, Q9WU53, Q9WU54 | Gene names | Birc5, Api4, Iap4 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 5 (Apoptosis inhibitor survivin) (Apoptosis inhibitor 4) (TIAP). | |||||
|
CD14_MOUSE
|
||||||
| NC score | 0.050947 (rank : 55) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10810 | Gene names | Cd14 | |||
|
Domain Architecture |

|
|||||
| Description | Monocyte differentiation antigen CD14 precursor (Myeloid cell-specific leucine-rich glycoprotein). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.029716 (rank : 56) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
PEO1_HUMAN
|
||||||
| NC score | 0.029406 (rank : 57) | θ value | 0.62314 (rank : 38) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96RR1, Q6MZX2, Q96RR0 | Gene names | PEO1, C10orf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein). | |||||
|
PEO1_MOUSE
|
||||||
| NC score | 0.026117 (rank : 58) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CIW5, Q8K1Z1 | Gene names | Peo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Twinkle protein, mitochondrial precursor (EC 3.6.1.-) (T7 gp4-like protein with intramitochondrial nucleoid localization) (T7-like mitochondrial DNA helicase) (Progressive external ophthalmoplegia 1 protein homolog). | |||||
|
TRP13_MOUSE
|
||||||
| NC score | 0.025003 (rank : 59) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q3UA06, Q3UQG6, Q9CWW8 | Gene names | Trip13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13). | |||||
|
TRP13_HUMAN
|
||||||
| NC score | 0.023737 (rank : 60) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q15645, O15324 | Gene names | TRIP13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 13 (Thyroid hormone receptor interactor 13) (Trip-13) (Human papillomavirus type 16 E1 protein- binding protein) (HPV16 E1 protein-binding protein) (16E1-BP). | |||||
|
CLPX_MOUSE
|
||||||
| NC score | 0.022344 (rank : 61) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JHS4, Q9WVD1 | Gene names | Clpx | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
BMS1_HUMAN
|
||||||
| NC score | 0.021475 (rank : 62) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14692, Q86XJ9 | Gene names | BMS1L, KIAA0187 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome biogenesis protein BMS1 homolog. | |||||
|
NVL_MOUSE
|
||||||
| NC score | 0.020852 (rank : 63) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9DBY8, Q3USC4, Q8BW27, Q8K2B5 | Gene names | Nvl | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
PEX6_HUMAN
|
||||||
| NC score | 0.020078 (rank : 64) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13608, Q8WYQ2, Q99476 | Gene names | PEX6, PXAAA1 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
PEX6_MOUSE
|
||||||
| NC score | 0.020059 (rank : 65) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99LC9, Q6YNQ9 | Gene names | Pex6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome assembly factor 2 (PAF-2) (Peroxisomal-type ATPase 1) (Peroxin-6) (Peroxisomal biogenesis factor 6). | |||||
|
NVL_HUMAN
|
||||||
| NC score | 0.016579 (rank : 66) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O15381, Q96EM7 | Gene names | NVL | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear valosin-containing protein-like (Nuclear VCP-like protein) (NVLp). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.015981 (rank : 67) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
PNPO_MOUSE
|
||||||
| NC score | 0.015712 (rank : 68) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91XF0 | Gene names | Pnpo | |||
|
Domain Architecture |

|
|||||
| Description | Pyridoxine-5'-phosphate oxidase (EC 1.4.3.5) (Pyridoxamine-phosphate oxidase). | |||||
|
VPS4B_MOUSE
|
||||||
| NC score | 0.011314 (rank : 69) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P46467, Q91W22, Q9R1C9 | Gene names | Vps4b, Skd1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associating protein 4B (Protein SKD1). | |||||
|
PRS10_HUMAN
|
||||||
| NC score | 0.010805 (rank : 70) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62333, P49719, Q6IBU3, Q92524 | Gene names | PSMC6, SUG2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
PRS10_MOUSE
|
||||||
| NC score | 0.010805 (rank : 71) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62334, P49719, Q3TKK1, Q810A6, Q92524, Q9CXH9 | Gene names | Psmc6, Sug2 | |||
|
Domain Architecture |

|
|||||
| Description | 26S protease regulatory subunit S10B (Proteasome subunit p42) (Proteasome 26S subunit ATPase 6). | |||||
|
KGUA_MOUSE
|
||||||
| NC score | 0.010065 (rank : 72) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64520 | Gene names | Guk1, Gmk | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate kinase (EC 2.7.4.8) (GMP kinase). | |||||
|
DIRA3_HUMAN
|
||||||
| NC score | 0.007173 (rank : 73) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95661 | Gene names | DIRAS3, ARHI, NOEY2, RHOI | |||
|
Domain Architecture |

|
|||||
| Description | GTP-binding protein Di-Ras3 (Distinct subgroup of the Ras family member 3) (Rho-related GTP-binding protein RhoI). | |||||
|
DHX57_HUMAN
|
||||||
| NC score | 0.006527 (rank : 74) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P158, Q53SI4, Q6P9G1, Q7Z6H3, Q8NG17, Q96M33 | Gene names | DHX57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
DHX57_MOUSE
|
||||||
| NC score | 0.006496 (rank : 75) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5D3, Q3TS93, Q6NZK4, Q6P1B4, Q8BI63, Q8BIA2, Q8R360 | Gene names | Dhx57 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative ATP-dependent RNA helicase DHX57 (EC 3.6.1.-) (DEAH box protein 57). | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.005364 (rank : 76) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.001893 (rank : 77) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.001339 (rank : 78) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
LRIG3_MOUSE
|
||||||
| NC score | -0.000680 (rank : 79) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P1C6, Q6ZPE6, Q8BRF0, Q8BS51, Q8C5E4 | Gene names | Lrig3, Kiaa3016 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 3 precursor. | |||||
|
ZNF35_HUMAN
|
||||||
| NC score | -0.003495 (rank : 80) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 65 | Target Neighborhood Hits | 761 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13682, Q96D01 | Gene names | ZNF35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 35 (Zinc finger protein HF.10). | |||||