Please be patient as the page loads
|
NALP6_MOUSE
|
||||||
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NALP6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993117 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
CIAS1_HUMAN
|
||||||
| θ value | 6.25694e-102 (rank : 3) | NC score | 0.948892 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| θ value | 1.44246e-98 (rank : 4) | NC score | 0.947681 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL12_HUMAN
|
||||||
| θ value | 1.88391e-98 (rank : 5) | NC score | 0.944673 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NAL14_HUMAN
|
||||||
| θ value | 4.82422e-78 (rank : 6) | NC score | 0.930973 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP1_HUMAN
|
||||||
| θ value | 1.22964e-73 (rank : 7) | NC score | 0.910068 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP4_HUMAN
|
||||||
| θ value | 1.60597e-73 (rank : 8) | NC score | 0.934200 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NALP2_HUMAN
|
||||||
| θ value | 1.96316e-71 (rank : 9) | NC score | 0.930387 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 2.17557e-62 (rank : 10) | NC score | 0.908555 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP7_HUMAN
|
||||||
| θ value | 1.84173e-61 (rank : 11) | NC score | 0.934865 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NAL10_HUMAN
|
||||||
| θ value | 2.48917e-58 (rank : 12) | NC score | 0.929837 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| θ value | 4.24587e-58 (rank : 13) | NC score | 0.926400 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 1.78386e-56 (rank : 14) | NC score | 0.917306 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 1.19655e-52 (rank : 15) | NC score | 0.843119 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
NAL11_HUMAN
|
||||||
| θ value | 9.18288e-45 (rank : 16) | NC score | 0.899936 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
C2TA_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 17) | NC score | 0.665727 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CAR15_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 18) | NC score | 0.698672 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
RINI_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 19) | NC score | 0.779237 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| θ value | 4.44505e-15 (rank : 20) | NC score | 0.768782 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
CARD4_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 21) | NC score | 0.667781 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
CAR15_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 22) | NC score | 0.713553 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
C2TA_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 23) | NC score | 0.634721 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CARD4_HUMAN
|
||||||
| θ value | 3.76295e-14 (rank : 24) | NC score | 0.659385 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
ASC_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 25) | NC score | 0.328139 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |
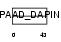
|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
ASC_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 26) | NC score | 0.384596 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
BIR1F_MOUSE
|
||||||
| θ value | 7.1131e-05 (rank : 27) | NC score | 0.234559 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1A_MOUSE
|
||||||
| θ value | 9.29e-05 (rank : 28) | NC score | 0.239591 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1E_MOUSE
|
||||||
| θ value | 0.000158464 (rank : 29) | NC score | 0.232699 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 30) | NC score | 0.225984 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
MEFV_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 31) | NC score | 0.176681 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
BIRC1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 32) | NC score | 0.255729 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
BIR1B_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 33) | NC score | 0.216269 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
DYH9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 34) | NC score | 0.022384 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
MYLIP_MOUSE
|
||||||
| θ value | 0.279714 (rank : 35) | NC score | 0.014802 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
CLPX_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.030041 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
VPP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.019119 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15920, Q3U2X3, Q9JHJ2 | Gene names | Atp6v0a2, Atp6n1b, Tj6 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (Immune suppressor factor J6B7). | |||||
|
LRC34_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.439672 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
RASA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.009415 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
TMOD2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.051090 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JKK7, Q9JLH9 | Gene names | Tmod2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
VPP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.018590 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y487 | Gene names | ATP6V0A2 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (TJ6). | |||||
|
ABTB2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.011062 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
CAR12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.310367 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
CF154_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.413047 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC34_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.399470 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.009661 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
TMOD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.056333 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZR1 | Gene names | TMOD2, NTMOD | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
COE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.010219 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.010219 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
ACLY_MOUSE
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.009538 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
ETS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.003506 (rank : 63) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
HDAC7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.004287 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
LIPB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.002933 (rank : 64) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.009228 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.004722 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
BIRC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.050842 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
CARD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.126744 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
LRC14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.073139 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
LRC31_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.398295 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.122938 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
LRC45_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.132105 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MEFV_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.089046 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
RGP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.263275 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
RGP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.251852 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
NALP6_HUMAN
|
||||||
| NC score | 0.993117 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P59044 | Gene names | NALP6, PYPAF5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5). | |||||
|
CIAS1_HUMAN
|
||||||
| NC score | 0.948892 (rank : 3) | θ value | 6.25694e-102 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96P20, O75434, Q59H68, Q5JQS8, Q5JQS9, Q6TG35, Q8TCW0, Q8TEU9, Q8WXH9 | Gene names | CIAS1, NALP3, PYPAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein (Cryopyrin) (NACHT-, LRR- and PYD-containing protein 3) (PYRIN-containing APAF1-like protein 1) (Angiotensin/vasopressin receptor AII/AVP-like). | |||||
|
CIAS1_MOUSE
|
||||||
| NC score | 0.947681 (rank : 4) | θ value | 1.44246e-98 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8R4B8, Q6JEL0 | Gene names | Cias1, Mmig1, Nalp3, Pypaf1 | |||
|
Domain Architecture |

|
|||||
| Description | Cold autoinflammatory syndrome 1 protein homolog (PYRIN-containing APAF1-like protein 1) (Mast cell maturation-associated-inducible protein 1) (Cryopyrin). | |||||
|
NAL12_HUMAN
|
||||||
| NC score | 0.944673 (rank : 5) | θ value | 1.88391e-98 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P59046, Q8NEU4, Q9BY26 | Gene names | NALP12, PYPAF7, RNO | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 12 (PYRIN-containing APAF1-like protein 7) (Monarch-1) (Regulated by nitric oxide). | |||||
|
NALP7_HUMAN
|
||||||
| NC score | 0.934865 (rank : 6) | θ value | 1.84173e-61 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WX94, Q7RTR1 | Gene names | NALP7, NOD12, PYPAF3 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 7 (PYRIN-containing APAF1-like protein 3). | |||||
|
NALP4_HUMAN
|
||||||
| NC score | 0.934200 (rank : 7) | θ value | 1.60597e-73 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MN2, Q86W87, Q96AY6 | Gene names | NALP4, PAN2, PYPAF4, RNH2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 4 (PYRIN-containing APAF1-like protein 4) (PAAD and NACHT-containing protein 2) (PYRIN and NACHT- containing protein 2) (Ribonuclease inhibitor 2). | |||||
|
NAL14_HUMAN
|
||||||
| NC score | 0.930973 (rank : 8) | θ value | 4.82422e-78 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86W24, Q7RTR6 | Gene names | NALP14, NOD5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 14 (Nucleotide-binding oligomerization domain protein 5). | |||||
|
NALP2_HUMAN
|
||||||
| NC score | 0.930387 (rank : 9) | θ value | 1.96316e-71 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9NX02, Q53FL5, Q59G09, Q8IXT0, Q9BVN5, Q9H6G6, Q9HAV9, Q9NWK3 | Gene names | NALP2, NBS1, PAN1, PYPAF2 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 2 (PYRIN domain and NACHT domain-containing protein 1) (PYRIN-containing APAF1-like protein 2) (Nucleotide-binding site protein 1). | |||||
|
NAL10_HUMAN
|
||||||
| NC score | 0.929837 (rank : 10) | θ value | 2.48917e-58 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86W26, Q6JGT0 | Gene names | NALP10, NOD8, PYNOD | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10 (Nucleotide-binding oligomerization domain protein 8). | |||||
|
NAL10_MOUSE
|
||||||
| NC score | 0.926400 (rank : 11) | θ value | 4.24587e-58 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8CCN1 | Gene names | Nalp10, Pynod | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 10. | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.917306 (rank : 12) | θ value | 1.78386e-56 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
NALP1_HUMAN
|
||||||
| NC score | 0.910068 (rank : 13) | θ value | 1.22964e-73 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9C000, Q9BZZ8, Q9BZZ9, Q9HAV8, Q9UFT4, Q9Y2E0 | Gene names | NALP1, CARD7, DEFCAP, KIAA0926, NAC | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 1 (Death effector filament- forming ced-4-like apoptosis protein) (Nucleotide-binding domain and caspase recruitment domain) (Caspase recruitment domain protein 7). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.908555 (rank : 14) | θ value | 2.17557e-62 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NAL11_HUMAN
|
||||||
| NC score | 0.899936 (rank : 15) | θ value | 9.18288e-45 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P59045, Q53ZZ0, Q8NBF5 | Gene names | NALP11, NOD17, PYPAF6 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 11 (PYRIN-containing APAF1-like protein 6) (Nucleotide-binding oligomerization domain protein 17). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.843119 (rank : 16) | θ value | 1.19655e-52 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
RINI_HUMAN
|
||||||
| NC score | 0.779237 (rank : 17) | θ value | 2.13179e-17 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P13489 | Gene names | RNH1, PRI, RNH | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1) (RAI) (Placental ribonuclease inhibitor) (RNase inhibitor) (RI). | |||||
|
RINI_MOUSE
|
||||||
| NC score | 0.768782 (rank : 18) | θ value | 4.44505e-15 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91VI7, Q924P4 | Gene names | Rnh1, Rnh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribonuclease inhibitor (Ribonuclease/angiogenin inhibitor 1). | |||||
|
CAR15_MOUSE
|
||||||
| NC score | 0.713553 (rank : 19) | θ value | 2.20605e-14 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K3Z0 | Gene names | Card15, Nod2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2). | |||||
|
CAR15_HUMAN
|
||||||
| NC score | 0.698672 (rank : 20) | θ value | 2.13179e-17 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9HC29, Q96RH5, Q96RH6, Q96RH8 | Gene names | CARD15, IBD1, NOD2 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 15 (Protein Nod2) (Inflammatory bowel disease protein 1). | |||||
|
CARD4_MOUSE
|
||||||
| NC score | 0.667781 (rank : 21) | θ value | 1.29331e-14 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BHB0, Q8BUT6 | Gene names | Card4 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4. | |||||
|
C2TA_MOUSE
|
||||||
| NC score | 0.665727 (rank : 22) | θ value | 2.06002e-20 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P79621, O46787, O78036, O78109, Q31115, Q9TPP1 | Gene names | Ciita, C2ta, Mhc2ta | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
CARD4_HUMAN
|
||||||
| NC score | 0.659385 (rank : 23) | θ value | 3.76295e-14 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y239, Q8IWF5 | Gene names | CARD4, NOD1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 4 (Protein Nod1). | |||||
|
C2TA_HUMAN
|
||||||
| NC score | 0.634721 (rank : 24) | θ value | 3.76295e-14 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P33076 | Gene names | CIITA, MHC2TA | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II transactivator (CIITA). | |||||
|
LRC34_HUMAN
|
||||||
| NC score | 0.439672 (rank : 25) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IZ02 | Gene names | LRRC34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
CF154_HUMAN
|
||||||
| NC score | 0.413047 (rank : 26) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5JTD7 | Gene names | C6orf154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf154. | |||||
|
LRC34_MOUSE
|
||||||
| NC score | 0.399470 (rank : 27) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9DAM1 | Gene names | Lrrc34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 34. | |||||
|
LRC31_HUMAN
|
||||||
| NC score | 0.398295 (rank : 28) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6UY01, Q8N848, Q9H5N5 | Gene names | LRRC31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 31. | |||||
|
ASC_MOUSE
|
||||||
| NC score | 0.384596 (rank : 29) | θ value | 6.43352e-06 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9EPB4, Q9D2W9 | Gene names | Pycard, Asc | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (mASC) (PYD and CARD domain-containing protein). | |||||
|
ASC_HUMAN
|
||||||
| NC score | 0.328139 (rank : 30) | θ value | 4.45536e-07 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULZ3, Q96D12, Q9BSZ5, Q9HBD0, Q9NXJ8 | Gene names | PYCARD, ASC, CARD5, TMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis-associated speck-like protein containing a CARD (hASC) (PYD and CARD domain-containing protein) (Target of methylation-induced silencing 1) (Caspase recruitment domain-containing protein 5). | |||||
|
CAR12_HUMAN
|
||||||
| NC score | 0.310367 (rank : 31) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NPP4, Q96J81, Q96J82, Q96J83 | Gene names | CARD12, CLAN, CLAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 12 (Ice protease- activating factor) (Ipaf) (CARD, LRR, and NACHT-containing protein) (Clan protein). | |||||
|
RGP1_HUMAN
|
||||||
| NC score | 0.263275 (rank : 32) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P46060 | Gene names | RANGAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
BIRC1_HUMAN
|
||||||
| NC score | 0.255729 (rank : 33) | θ value | 0.00509761 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13075, O75857, Q13730, Q99796 | Gene names | BIRC1, NAIP | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1 (Neuronal apoptosis inhibitory protein). | |||||
|
RGP1_MOUSE
|
||||||
| NC score | 0.251852 (rank : 34) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P46061, Q60801 | Gene names | Rangap1, Fug1 | |||
|
Domain Architecture |

|
|||||
| Description | Ran GTPase-activating protein 1. | |||||
|
BIR1A_MOUSE
|
||||||
| NC score | 0.239591 (rank : 35) | θ value | 9.29e-05 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9QWK5, Q9JIB5, Q9R017 | Gene names | Birc1a, Naip, Naip1 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1a (Neuronal apoptosis inhibitory protein 1). | |||||
|
BIR1F_MOUSE
|
||||||
| NC score | 0.234559 (rank : 36) | θ value | 7.1131e-05 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JIB6, O09121, O09122, P81704 | Gene names | Birc1f, Naip-rs4, Naip6 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1f (Neuronal apoptosis inhibitory protein 6). | |||||
|
BIR1E_MOUSE
|
||||||
| NC score | 0.232699 (rank : 37) | θ value | 0.000158464 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9R016, O09121, O09122, P81703, Q9R029 | Gene names | Birc1e, Naip-rs3, Naip5 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1e (Neuronal apoptosis inhibitory protein 5). | |||||
|
BIR1G_MOUSE
|
||||||
| NC score | 0.225984 (rank : 38) | θ value | 0.00035302 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9JIB3 | Gene names | Birc1g, Naip7 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1g (Neuronal apoptosis inhibitory protein 7). | |||||
|
BIR1B_MOUSE
|
||||||
| NC score | 0.216269 (rank : 39) | θ value | 0.0252991 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9QUK4, O09124, Q9R030 | Gene names | Birc1b, Naip-rs6, Naip2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 1b (Neuronal apoptosis inhibitory protein 2). | |||||
|
MEFV_MOUSE
|
||||||
| NC score | 0.176681 (rank : 40) | θ value | 0.00134147 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JJ26 | Gene names | Mefv | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC45_MOUSE
|
||||||
| NC score | 0.132105 (rank : 41) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CIM1, Q3U5Z2 | Gene names | Lrrc45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
CARD8_HUMAN
|
||||||
| NC score | 0.126744 (rank : 42) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2G2, Q6PGP8, Q96P82 | Gene names | CARD8, KIAA0955, NDPP1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 8 (Apoptotic protein NDPP1) (DACAR) (CARD-inhibitor of NF-kappa-B-activating ligand) (CARDINAL) (Tumor-up-regulated CARD-containing antagonist of CASP9) (TUCAN). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.122938 (rank : 43) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.089046 (rank : 44) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
LRC14_HUMAN
|
||||||
| NC score | 0.073139 (rank : 45) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q15048 | Gene names | LRRC14, KIAA0014 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat-containing protein 14. | |||||
|
TMOD2_HUMAN
|
||||||
| NC score | 0.056333 (rank : 46) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZR1 | Gene names | TMOD2, NTMOD | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
TMOD2_MOUSE
|
||||||
| NC score | 0.051090 (rank : 47) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9JKK7, Q9JLH9 | Gene names | Tmod2 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomodulin-2 (Neuronal tropomodulin) (N-Tmod). | |||||
|
BIRC2_MOUSE
|
||||||
| NC score | 0.050842 (rank : 48) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62210, O08864 | Gene names | Birc2, Birc3, Iap2 | |||
|
Domain Architecture |

|
|||||
| Description | Baculoviral IAP repeat-containing protein 2 (Inhibitor of apoptosis protein 2) (MIAP2) (MIAP-2). | |||||
|
CLPX_HUMAN
|
||||||
| NC score | 0.030041 (rank : 49) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76031, Q9H4D9 | Gene names | CLPX | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent Clp protease ATP-binding subunit ClpX-like, mitochondrial precursor. | |||||
|
DYH9_HUMAN
|
||||||
| NC score | 0.022384 (rank : 50) | θ value | 0.0736092 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NYC9, O95494, Q9NQ28 | Gene names | DNAH9, DNAH17L, DNAL1 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 9 (Axonemal beta dynein heavy chain 9). | |||||
|
VPP2_MOUSE
|
||||||
| NC score | 0.019119 (rank : 51) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15920, Q3U2X3, Q9JHJ2 | Gene names | Atp6v0a2, Atp6n1b, Tj6 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (Immune suppressor factor J6B7). | |||||
|
VPP2_HUMAN
|
||||||
| NC score | 0.018590 (rank : 52) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y487 | Gene names | ATP6V0A2 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 2 (V- ATPase 116 kDa isoform a2) (TJ6). | |||||
|
MYLIP_MOUSE
|
||||||
| NC score | 0.014802 (rank : 53) | θ value | 0.279714 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BM54, Q3TX01, Q8BLY9, Q91Z47 | Gene names | Mylip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase MYLIP (EC 6.3.2.-) (Myosin regulatory light chain- interacting protein) (MIR). | |||||
|
ABTB2_MOUSE
|
||||||
| NC score | 0.011062 (rank : 54) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
COE1_HUMAN
|
||||||
| NC score | 0.010219 (rank : 55) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| NC score | 0.010219 (rank : 56) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.009661 (rank : 57) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ACLY_MOUSE
|
||||||
| NC score | 0.009538 (rank : 58) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
RASA2_HUMAN
|
||||||
| NC score | 0.009415 (rank : 59) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15283, O00695, Q15284, Q92594, Q99577, Q9UEQ2 | Gene names | RASA2, GAP1M, RASGAP | |||
|
Domain Architecture |

|
|||||
| Description | Ras GTPase-activating protein 2 (GAP1m). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.009228 (rank : 60) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.004722 (rank : 61) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
HDAC7_MOUSE
|
||||||
| NC score | 0.004287 (rank : 62) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
ETS1_MOUSE
|
||||||
| NC score | 0.003506 (rank : 63) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27577, Q61403 | Gene names | Ets1, Ets-1 | |||
|
Domain Architecture |

|
|||||
| Description | C-ets-1 protein (p54). | |||||
|
LIPB1_HUMAN
|
||||||
| NC score | 0.002933 (rank : 64) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 640 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W92, O75336, Q86X70, Q9NY03, Q9ULJ0 | Gene names | PPFIBP1, KIAA1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1) (hSGT2). | |||||