Please be patient as the page loads
|
ACLY_MOUSE
|
||||||
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
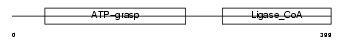
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ACLY_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999350 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53396, Q13037, Q9BRL0 | Gene names | ACLY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
ACLY_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
SUCA_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 3) | NC score | 0.599512 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUM5, Q8C2C3, Q9DBA3, Q9DCI8 | Gene names | Suclg1 | |||
|
Domain Architecture |
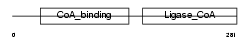
|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
SUCA_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 4) | NC score | 0.582419 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53597, Q9BWB0, Q9UNP6 | Gene names | SUCLG1 | |||
|
Domain Architecture |
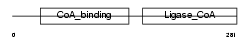
|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
SUCB2_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 5) | NC score | 0.349971 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I8, Q3T9B8, Q3TJQ5, Q3TK63, Q66JT3, Q7TMY3, Q80VV1, Q8K2K9 | Gene names | Suclg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP- specific succinyl-CoA synthetase subunit beta). | |||||
|
SUCB2_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 6) | NC score | 0.343081 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I99, O95195, Q6NUH7, Q86VX8, Q8WUQ1 | Gene names | SUCLG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP- specific succinyl-CoA synthetase subunit beta). | |||||
|
SUCB1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 7) | NC score | 0.329696 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I9, Q3TVH1, Q8BGS6 | Gene names | Sucla2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.5) (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP- specific succinyl-CoA synthetase subunit beta). | |||||
|
SUCB1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 8) | NC score | 0.318234 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2R7, O95194, Q9NV21, Q9NVP7 | Gene names | SUCLA2 | |||
|
Domain Architecture |
|
|||||
| Description | Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.5) (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP- specific succinyl-CoA synthetase subunit beta) (NY-REN-39 antigen). | |||||
|
CACP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.044803 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43155, Q9BW16 | Gene names | CRAT, CAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Carnitine O-acetyltransferase (EC 2.3.1.7) (Carnitine acetylase) (CAT) (Carnitine acetyltransferase) (CrAT). | |||||
|
SP3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.018072 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.026018 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.026045 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SP3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.015462 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70494, Q68FF2, Q8CF64, Q8K378 | Gene names | Sp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp3. | |||||
|
BMP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.010976 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22003, Q9H547, Q9NTM5 | Gene names | BMP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 5 precursor (BMP-5). | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.019699 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
NALP6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.009538 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
DHRS4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.008321 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
GRIP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.008454 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
PPBT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.014385 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05186, Q8WU32, Q9UBK0 | Gene names | ALPL | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phosphatase, tissue-nonspecific isozyme precursor (EC 3.1.3.1) (AP-TNAP) (Liver/bone/kidney isozyme) (TNSALP). | |||||
|
KSYK_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.001048 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
PDZD2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.006516 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
ACLY_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
ACLY_HUMAN
|
||||||
| NC score | 0.999350 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P53396, Q13037, Q9BRL0 | Gene names | ACLY | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
SUCA_MOUSE
|
||||||
| NC score | 0.599512 (rank : 3) | θ value | 2.97466e-19 (rank : 3) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WUM5, Q8C2C3, Q9DBA3, Q9DCI8 | Gene names | Suclg1 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
SUCA_HUMAN
|
||||||
| NC score | 0.582419 (rank : 4) | θ value | 2.7842e-17 (rank : 4) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53597, Q9BWB0, Q9UNP6 | Gene names | SUCLG1 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [GDP-forming] subunit alpha, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase subunit alpha) (SCS- alpha). | |||||
|
SUCB2_MOUSE
|
||||||
| NC score | 0.349971 (rank : 5) | θ value | 4.0297e-08 (rank : 5) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I8, Q3T9B8, Q3TJQ5, Q3TK63, Q66JT3, Q7TMY3, Q80VV1, Q8K2K9 | Gene names | Suclg2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP- specific succinyl-CoA synthetase subunit beta). | |||||
|
SUCB2_HUMAN
|
||||||
| NC score | 0.343081 (rank : 6) | θ value | 1.53129e-07 (rank : 6) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96I99, O95195, Q6NUH7, Q86VX8, Q8WUQ1 | Gene names | SUCLG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.4) (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP- specific succinyl-CoA synthetase subunit beta). | |||||
|
SUCB1_MOUSE
|
||||||
| NC score | 0.329696 (rank : 7) | θ value | 1.69304e-06 (rank : 7) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2I9, Q3TVH1, Q8BGS6 | Gene names | Sucla2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.5) (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP- specific succinyl-CoA synthetase subunit beta). | |||||
|
SUCB1_HUMAN
|
||||||
| NC score | 0.318234 (rank : 8) | θ value | 8.40245e-06 (rank : 8) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P2R7, O95194, Q9NV21, Q9NVP7 | Gene names | SUCLA2 | |||
|
Domain Architecture |

|
|||||
| Description | Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (EC 6.2.1.5) (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP- specific succinyl-CoA synthetase subunit beta) (NY-REN-39 antigen). | |||||
|
CACP_HUMAN
|
||||||
| NC score | 0.044803 (rank : 9) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P43155, Q9BW16 | Gene names | CRAT, CAT1 | |||
|
Domain Architecture |

|
|||||
| Description | Carnitine O-acetyltransferase (EC 2.3.1.7) (Carnitine acetylase) (CAT) (Carnitine acetyltransferase) (CrAT). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.026045 (rank : 10) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.026018 (rank : 11) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.019699 (rank : 12) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
SP3_HUMAN
|
||||||
| NC score | 0.018072 (rank : 13) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02447, Q8TD56, Q9BQR1 | Gene names | SP3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp3 (SPR-2). | |||||
|
SP3_MOUSE
|
||||||
| NC score | 0.015462 (rank : 14) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 731 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70494, Q68FF2, Q8CF64, Q8K378 | Gene names | Sp3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp3. | |||||
|
PPBT_HUMAN
|
||||||
| NC score | 0.014385 (rank : 15) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P05186, Q8WU32, Q9UBK0 | Gene names | ALPL | |||
|
Domain Architecture |

|
|||||
| Description | Alkaline phosphatase, tissue-nonspecific isozyme precursor (EC 3.1.3.1) (AP-TNAP) (Liver/bone/kidney isozyme) (TNSALP). | |||||
|
BMP5_HUMAN
|
||||||
| NC score | 0.010976 (rank : 16) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22003, Q9H547, Q9NTM5 | Gene names | BMP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 5 precursor (BMP-5). | |||||
|
NALP6_MOUSE
|
||||||
| NC score | 0.009538 (rank : 17) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91WS2, Q8K0L4 | Gene names | Nalp6, Pypaf5 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 6 (PYRIN-containing APAF1-like protein 5-like). | |||||
|
GRIP2_HUMAN
|
||||||
| NC score | 0.008454 (rank : 18) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9C0E4, Q8TEH9, Q9H7H3 | Gene names | GRIP2, KIAA1719 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 2 (GRIP2 protein). | |||||
|
DHRS4_HUMAN
|
||||||
| NC score | 0.008321 (rank : 19) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTZ2, O95162, Q6UWU3, Q8TD03, Q9H3N5, Q9NV08 | Gene names | DHRS4 | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrogenase/reductase SDR family member 4 (EC 1.1.1.184) (NADPH- dependent carbonyl reductase/NADP-retinol dehydrogenase) (CR) (PHCR) (Peroxisomal short-chain alcohol dehydrogenase) (NADPH-dependent retinol dehydrogenase/reductase) (NDRD) (SCAD-SRL) (humNRDR) (PSCD). | |||||
|
PDZD2_HUMAN
|
||||||
| NC score | 0.006516 (rank : 20) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15018, Q9BXD4 | Gene names | PDZD2, AIPC, KIAA0300, PDZK3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing protein 2 (PDZ domain-containing protein 3) (Activated in prostate cancer protein). | |||||
|
KSYK_MOUSE
|
||||||
| NC score | 0.001048 (rank : 21) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 21 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||