Please be patient as the page loads
|
HPS4_MOUSE
|
||||||
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HPS4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.954070 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQG7, Q5H8V6, Q96LX6, Q9BY93, Q9UH37, Q9UH38 | Gene names | HPS4, KIAA1667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein (Light-ear protein homolog). | |||||
|
HPS4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 3) | NC score | 0.025960 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
TATD2_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.098395 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
GLI2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.016294 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
EFHB_MOUSE
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.063415 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.047658 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.043469 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.037849 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.038174 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
EVL_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.041393 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
CEP68_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.055594 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
CJ047_MOUSE
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.061235 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.044439 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.019690 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.059658 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
WNK1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.024254 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.020961 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.055743 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.055715 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
ATS20_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.013567 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
CD45_MOUSE
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.014184 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
EOMES_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.018702 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |
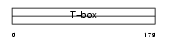
|
|||||
| Description | Eomesodermin homolog. | |||||
|
CAC1I_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.014951 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
GLIS1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.009743 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1M4, Q6NZF6, Q8R4Z3 | Gene names | Glis1, Gli5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1) (Gli homologous protein 1) (GliH1). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.051644 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PRR11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.042948 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHE0, Q3V3H1 | Gene names | Prr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.032980 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.038666 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FOXC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.017321 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |
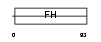
|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
GCM1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.029897 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |
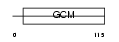
|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
|
LRFN5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.012680 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NI6, Q86XL2 | Gene names | LRFN5, C14orf146 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.035547 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
BAI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.012292 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.040289 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.017146 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
LRFN5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.012109 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.040514 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.032678 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.021166 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
DOCK4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.016520 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
FOXC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.015083 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.018756 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
ZN683_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.006137 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZ20, Q5T141, Q5T146, Q5T147, Q5T149, Q8NEN4 | Gene names | ZNF683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 683. | |||||
|
ACLY_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.019699 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
ARRS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.014082 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |
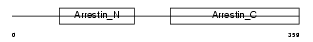
|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
DEN1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.025541 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.022707 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
FBX7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.031118 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
HES2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.022339 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
IASPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.018236 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IDUA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.022969 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48441 | Gene names | Idua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-L-iduronidase precursor (EC 3.2.1.76). | |||||
|
KIF12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.007530 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FN5, Q5TBE0 | Gene names | KIF12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
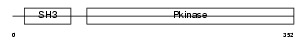
M3K10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.005178 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
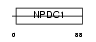
NPDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.026631 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQX5, Q9BTD6, Q9BXT3, Q9NQS2, Q9Y434 | Gene names | NPDC1 | |||
|
Domain Architecture |
|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.021871 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PXK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.025734 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.012649 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
SYNP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.020713 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
ZHX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.015178 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.015507 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DEN1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.023354 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.027532 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
DUX4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.012304 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBX2 | Gene names | DUX4, DUX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double homeobox protein 4 (Double homeobox protein 10) (Double homeobox protein 4/10). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.016567 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
LRMP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.019035 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
MYCBP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.027684 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQS3, O88308 | Gene names | Mycbp, Amy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.033856 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
U689_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.026368 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UX39 | Gene names | ames=UNQ689/PRO1329 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 precursor. | |||||
|
ZMYM3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.014387 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.004595 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN710_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.003346 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.037353 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ELL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.014091 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.006333 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
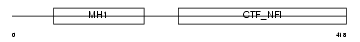
NFIX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.008960 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14938, O60413, Q12859, Q13050, Q13052, Q9UPH1, Q9Y6R8 | Gene names | NFIX | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear factor 1 X-type (Nuclear factor 1/X) (NF1-X) (NFI-X) (NF-I/X) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.023898 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SEM6D_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.009508 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.009420 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SIX5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.015078 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70178 | Gene names | Six5, Dmahp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein homolog). | |||||
|
HPS4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q99KG7 | Gene names | Hps4, Le | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein homolog (Light-ear protein) (Le protein). | |||||
|
HPS4_HUMAN
|
||||||
| NC score | 0.954070 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NQG7, Q5H8V6, Q96LX6, Q9BY93, Q9UH37, Q9UH38 | Gene names | HPS4, KIAA1667 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hermansky-Pudlak syndrome 4 protein (Light-ear protein homolog). | |||||
|
TATD2_HUMAN
|
||||||
| NC score | 0.098395 (rank : 3) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q93075 | Gene names | TATDN2, KIAA0218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TatD DNase domain-containing deoxyribonuclease 2 (EC 3.1.21.-). | |||||
|
EFHB_MOUSE
|
||||||
| NC score | 0.063415 (rank : 4) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8CDU5 | Gene names | Efhb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand domain-containing family member B. | |||||
|
CJ047_MOUSE
|
||||||
| NC score | 0.061235 (rank : 5) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C5R2, Q8R017, Q8R2Z9 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf47 homolog. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.059658 (rank : 6) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.055743 (rank : 7) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.055715 (rank : 8) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
CEP68_HUMAN
|
||||||
| NC score | 0.055594 (rank : 9) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76N32, O60326, Q9BQ18, Q9UDM9 | Gene names | CEP68, KIAA0582 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 68 kDa (Cep68 protein). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.051644 (rank : 10) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.047658 (rank : 11) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.044439 (rank : 12) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.043469 (rank : 13) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
PRR11_MOUSE
|
||||||
| NC score | 0.042948 (rank : 14) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BHE0, Q3V3H1 | Gene names | Prr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
EVL_MOUSE
|
||||||
| NC score | 0.041393 (rank : 15) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70429, Q9ERU8 | Gene names | Evl | |||
|
Domain Architecture |

|
|||||
| Description | Ena/VASP-like protein (Ena/vasodilator-stimulated phosphoprotein- like). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.040514 (rank : 16) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.040289 (rank : 17) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.038666 (rank : 18) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.038174 (rank : 19) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.037849 (rank : 20) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.037353 (rank : 21) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.035547 (rank : 22) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.033856 (rank : 23) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.032980 (rank : 24) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.032678 (rank : 25) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
FBX7_HUMAN
|
||||||
| NC score | 0.031118 (rank : 26) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3I1, Q5TGC4, Q96HM6, Q9UF21, Q9UKT2 | Gene names | FBXO7, FBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 7. | |||||
|
GCM1_MOUSE
|
||||||
| NC score | 0.029897 (rank : 27) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70348, O09103 | Gene names | Gcm1, Gcma | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMa (Glial cells missing homolog 1) (GCM motif protein 1) (mGCMa) (mGCM1). | |||||
|
MYCBP_MOUSE
|
||||||
| NC score | 0.027684 (rank : 28) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9EQS3, O88308 | Gene names | Mycbp, Amy1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-Myc-binding protein (Associate of Myc 1) (AMY-1). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.027532 (rank : 29) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
NPDC1_HUMAN
|
||||||
| NC score | 0.026631 (rank : 30) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NQX5, Q9BTD6, Q9BXT3, Q9NQS2, Q9Y434 | Gene names | NPDC1 | |||
|
Domain Architecture |

|
|||||
| Description | Neural proliferation differentiation and control protein 1 precursor (NPDC-1 protein). | |||||
|
U689_HUMAN
|
||||||
| NC score | 0.026368 (rank : 31) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6UX39 | Gene names | ames=UNQ689/PRO1329 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein UNQ689/PRO1329 precursor. | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.025960 (rank : 32) | θ value | 0.0193708 (rank : 3) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
PXK_MOUSE
|
||||||
| NC score | 0.025734 (rank : 33) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BX57, Q3TD60, Q3TEL7, Q3TZZ1, Q3U197, Q3U773, Q3UCS5, Q4FBH7, Q91WB6 | Gene names | Pxk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
DEN1A_MOUSE
|
||||||
| NC score | 0.025541 (rank : 34) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K382 | Gene names | Dennd1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
WNK1_MOUSE
|
||||||
| NC score | 0.024254 (rank : 35) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1179 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P83741 | Gene names | Wnk1, Prkwnk1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.023898 (rank : 36) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
DEN1A_HUMAN
|
||||||
| NC score | 0.023354 (rank : 37) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEH3, Q5VWF0, Q8IVD6, Q9H796 | Gene names | DENND1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 1A. | |||||
|
IDUA_MOUSE
|
||||||
| NC score | 0.022969 (rank : 38) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P48441 | Gene names | Idua | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-L-iduronidase precursor (EC 3.2.1.76). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.022707 (rank : 39) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
HES2_HUMAN
|
||||||
| NC score | 0.022339 (rank : 40) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y543, Q96EN4, Q9Y542 | Gene names | HES2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor HES-2 (Hairy and enhancer of split 2). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.021871 (rank : 41) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.021166 (rank : 42) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.020961 (rank : 43) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
SYNP2_MOUSE
|
||||||
| NC score | 0.020713 (rank : 44) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YE8, Q8C592 | Gene names | Synpo2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin-2 (Myopodin). | |||||
|
ACLY_MOUSE
|
||||||
| NC score | 0.019699 (rank : 45) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91V92 | Gene names | Acly | |||
|
Domain Architecture |

|
|||||
| Description | ATP-citrate synthase (EC 2.3.3.8) (ATP-citrate (pro-S-)-lyase) (Citrate cleavage enzyme). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.019690 (rank : 46) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
LRMP_MOUSE
|
||||||
| NC score | 0.019035 (rank : 47) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60664, Q7TMU8 | Gene names | Lrmp, Jaw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphoid-restricted membrane protein (Protein Jaw1). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.018756 (rank : 48) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
EOMES_MOUSE
|
||||||
| NC score | 0.018702 (rank : 49) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54839, Q9QYG7 | Gene names | Eomes, Tbr2 | |||
|
Domain Architecture |

|
|||||
| Description | Eomesodermin homolog. | |||||
|
IASPP_HUMAN
|
||||||
| NC score | 0.018236 (rank : 50) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8WUF5, Q2PNZ9, Q5DU71, Q5I1X4, Q6P1R7, Q6PKF8, Q9Y290 | Gene names | PPP1R13L, IASPP, NKIP1, PPP1R13BL, RAI | |||
|
Domain Architecture |

|
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
FOXC1_HUMAN
|
||||||
| NC score | 0.017321 (rank : 51) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q12948, Q86UP7, Q9BYM1, Q9NUE5, Q9UDD0, Q9UP06 | Gene names | FOXC1, FKHL7, FREAC3 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.017146 (rank : 52) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.016567 (rank : 53) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
DOCK4_MOUSE
|
||||||
| NC score | 0.016520 (rank : 54) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P59764 | Gene names | Dock4, Kiaa0716 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 4. | |||||
|
GLI2_HUMAN
|
||||||
| NC score | 0.016294 (rank : 55) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 971 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10070, O60252, O60253, O60254, O60255, Q15590, Q15591 | Gene names | GLI2, THP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI2 (Tax helper protein). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.015507 (rank : 56) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
ZHX1_MOUSE
|
||||||
| NC score | 0.015178 (rank : 57) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70121, Q8BQ68, Q8C6T4, Q8CJG3 | Gene names | Zhx1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 1. | |||||
|
FOXC1_MOUSE
|
||||||
| NC score | 0.015083 (rank : 58) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61572, O88409, Q61582 | Gene names | Foxc1, Fkh1, Fkhl7, Freac3, Mf1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein C1 (Forkhead-related protein FKHL7) (Forkhead- related transcription factor 3) (FREAC-3) (Transcription factor FKH-1) (Mesoderm/mesenchyme forkhead 1) (MF-1). | |||||
|
SIX5_MOUSE
|
||||||
| NC score | 0.015078 (rank : 59) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70178 | Gene names | Six5, Dmahp | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX5 (DM locus-associated homeodomain protein homolog). | |||||
|
CAC1I_HUMAN
|
||||||
| NC score | 0.014951 (rank : 60) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0X4, O95504, Q5JZ88, Q7Z6S9, Q8NFX6, Q9NZC8, Q9UH15, Q9UH30, Q9ULU9, Q9UNE6 | Gene names | CACNA1I, KIAA1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1I (Voltage- gated calcium channel subunit alpha Cav3.3) (Ca(v)3.3). | |||||
|
ZMYM3_MOUSE
|
||||||
| NC score | 0.014387 (rank : 61) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JLM4, Q80U17 | Gene names | Zmym3, Kiaa0385, Zfp261, Znf261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261) (DXHXS6673E protein). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.014184 (rank : 62) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
ELL2_HUMAN
|
||||||
| NC score | 0.014091 (rank : 63) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00472 | Gene names | ELL2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL2. | |||||
|
ARRS_HUMAN
|
||||||
| NC score | 0.014082 (rank : 64) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P10523, Q53SV3, Q99858 | Gene names | SAG | |||
|
Domain Architecture |

|
|||||
| Description | S-arrestin (Retinal S-antigen) (48 kDa protein) (S-AG) (Rod photoreceptor arrestin). | |||||
|
ATS20_HUMAN
|
||||||
| NC score | 0.013567 (rank : 65) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59510 | Gene names | ADAMTS20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 20) (ADAM-TS 20) (ADAM-TS20). | |||||
|
LRFN5_HUMAN
|
||||||
| NC score | 0.012680 (rank : 66) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96NI6, Q86XL2 | Gene names | LRFN5, C14orf146 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.012649 (rank : 67) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
DUX4_HUMAN
|
||||||
| NC score | 0.012304 (rank : 68) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UBX2 | Gene names | DUX4, DUX10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Double homeobox protein 4 (Double homeobox protein 10) (Double homeobox protein 4/10). | |||||
|
BAI1_HUMAN
|
||||||
| NC score | 0.012292 (rank : 69) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14514 | Gene names | BAI1 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific angiogenesis inhibitor 1 precursor. | |||||
|
LRFN5_MOUSE
|
||||||
| NC score | 0.012109 (rank : 70) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BXA0, Q8BJH4, Q8BZL0 | Gene names | Lrfn5 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeat and fibronectin type-III domain-containing protein 5 precursor. | |||||
|
GLIS1_MOUSE
|
||||||
| NC score | 0.009743 (rank : 71) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 751 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8K1M4, Q6NZF6, Q8R4Z3 | Gene names | Glis1, Gli5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS1 (GLI-similar 1) (Gli homologous protein 1) (GliH1). | |||||
|
SEM6D_HUMAN
|
||||||
| NC score | 0.009508 (rank : 72) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NFY4, Q8NFY3, Q8NFY5, Q8NFY6, Q8NFY7, Q9P249 | Gene names | SEMA6D, KIAA1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
SEM6D_MOUSE
|
||||||
| NC score | 0.009420 (rank : 73) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q76KF0, Q76KF1, Q76KF2, Q76KF3, Q76KF4, Q80TD0 | Gene names | Sema6d, Kiaa1479 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Semaphorin-6D precursor. | |||||
|
NFIX_HUMAN
|
||||||
| NC score | 0.008960 (rank : 74) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14938, O60413, Q12859, Q13050, Q13052, Q9UPH1, Q9Y6R8 | Gene names | NFIX | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 X-type (Nuclear factor 1/X) (NF1-X) (NFI-X) (NF-I/X) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
KIF12_HUMAN
|
||||||
| NC score | 0.007530 (rank : 75) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96FN5, Q5TBE0 | Gene names | KIF12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF12. | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.006333 (rank : 76) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
ZN683_HUMAN
|
||||||
| NC score | 0.006137 (rank : 77) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZ20, Q5T141, Q5T146, Q5T147, Q5T149, Q8NEN4 | Gene names | ZNF683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 683. | |||||
|
M3K10_HUMAN
|
||||||
| NC score | 0.005178 (rank : 78) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q02779, Q12761, Q14871 | Gene names | MAP3K10, MLK2, MST | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 10 (EC 2.7.11.25) (Mixed lineage kinase 2) (Protein kinase MST). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.004595 (rank : 79) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN710_MOUSE
|
||||||
| NC score | 0.003346 (rank : 80) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 80 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q3U288, Q3U341, Q6P9R3, Q8BJH0, Q8BTL6 | Gene names | Znf710, Zfp710 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 710. | |||||