Please be patient as the page loads
|
DPOLA_MOUSE
|
||||||
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DPOLA_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990766 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09884, Q86UQ7 | Gene names | POLA1, POLA | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOLA_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOD1_MOUSE
|
||||||
| θ value | 3.60269e-49 (rank : 3) | NC score | 0.758246 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52431, O54883 | Gene names | Pold1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7). | |||||
|
DPOD1_HUMAN
|
||||||
| θ value | 3.37202e-47 (rank : 4) | NC score | 0.753661 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28340, Q8NER3, Q96H98 | Gene names | POLD1, POLD | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7) (DNA polymerase subunit delta p125). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 1.46948e-34 (rank : 5) | NC score | 0.638419 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 1.6247e-33 (rank : 6) | NC score | 0.639734 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
DPOE1_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 7) | NC score | 0.209954 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
DPOE1_MOUSE
|
||||||
| θ value | 0.000270298 (rank : 8) | NC score | 0.201230 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVF7, Q9QX50 | Gene names | Pole, Pole1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 9) | NC score | 0.071048 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.070306 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.059848 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
SPAT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.071602 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.029203 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.064287 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.028028 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
BMP2K_MOUSE
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.010057 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
CDN1C_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.039149 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.029072 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TNIK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.006224 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
DHX8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.012673 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
RAIN_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.040820 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U651, Q6U676 | Gene names | RASIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.023122 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.029211 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
ELF2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.030995 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
PPAC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.050948 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24666, P24667, Q16035, Q16036, Q16725 | Gene names | ACP1 | |||
|
Domain Architecture |
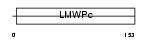
|
|||||
| Description | Low molecular weight phosphotyrosine protein phosphatase (EC 3.1.3.48) (LMW-PTP) (Low molecular weight cytosolic acid phosphatase) (EC 3.1.3.2) (Red cell acid phosphatase 1) (PTPase) (Adipocyte acid phosphatase). | |||||
|
AKAP8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.030783 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.003046 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.027982 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.005263 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
TTF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.016168 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
G3BP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.016789 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.013827 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NU160_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.030592 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12769, Q96GB3, Q9H660 | Gene names | NUP160, KIAA0197, NUP120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin). | |||||
|
NU160_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.035837 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0W3, Q3TBI7, Q3TP11, Q3U250, Q6A0A7, Q7TME1, Q9CZD9 | Gene names | Nup160, Gtl1-13, Kiaa0197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin) (Gene trap locus 1-13) (GTL-13). | |||||
|
ST1C1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.009608 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VR3, O70262 | Gene names | Sult1c1, Sult1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 1C1 (EC 2.8.2.-) (Phenol sulfotransferase). | |||||
|
T240L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.016919 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.013951 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
K1849_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.017215 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
NUF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.019811 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
TNR6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.015205 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25446, Q9DCQ1 | Gene names | Fas, Apt1, Tnfrsf6 | |||
|
Domain Architecture |
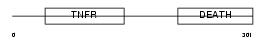
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
YTHD3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.020665 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z739, Q63Z37, Q659A3 | Gene names | YTHDF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
YTHD3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.020660 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
HCN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.006712 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.013500 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.003214 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ELF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.023064 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
MYT1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.011321 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.023258 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SMUF2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.004764 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.014669 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
CC47_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.024818 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
E41L2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.008466 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.009713 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.015042 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.008169 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.002167 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
DPOLA_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOLA_HUMAN
|
||||||
| NC score | 0.990766 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09884, Q86UQ7 | Gene names | POLA1, POLA | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOD1_MOUSE
|
||||||
| NC score | 0.758246 (rank : 3) | θ value | 3.60269e-49 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52431, O54883 | Gene names | Pold1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7). | |||||
|
DPOD1_HUMAN
|
||||||
| NC score | 0.753661 (rank : 4) | θ value | 3.37202e-47 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28340, Q8NER3, Q96H98 | Gene names | POLD1, POLD | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7) (DNA polymerase subunit delta p125). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.639734 (rank : 5) | θ value | 1.6247e-33 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.638419 (rank : 6) | θ value | 1.46948e-34 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
DPOE1_HUMAN
|
||||||
| NC score | 0.209954 (rank : 7) | θ value | 3.19293e-05 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
DPOE1_MOUSE
|
||||||
| NC score | 0.201230 (rank : 8) | θ value | 0.000270298 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVF7, Q9QX50 | Gene names | Pole, Pole1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
SPAT2_HUMAN
|
||||||
| NC score | 0.071602 (rank : 9) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UM82, O94857 | Gene names | SPATA2, KIAA0757, PD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spermatogenesis-associated protein 2 (Spermatogenesis-associated protein PD1). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.071048 (rank : 10) | θ value | 0.0330416 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.070306 (rank : 11) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.064287 (rank : 12) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.059848 (rank : 13) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
PPAC_HUMAN
|
||||||
| NC score | 0.050948 (rank : 14) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24666, P24667, Q16035, Q16036, Q16725 | Gene names | ACP1 | |||
|
Domain Architecture |

|
|||||
| Description | Low molecular weight phosphotyrosine protein phosphatase (EC 3.1.3.48) (LMW-PTP) (Low molecular weight cytosolic acid phosphatase) (EC 3.1.3.2) (Red cell acid phosphatase 1) (PTPase) (Adipocyte acid phosphatase). | |||||
|
RAIN_HUMAN
|
||||||
| NC score | 0.040820 (rank : 15) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U651, Q6U676 | Gene names | RASIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-interacting protein 1 (Rain). | |||||
|
CDN1C_MOUSE
|
||||||
| NC score | 0.039149 (rank : 16) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49919 | Gene names | Cdkn1c, Kip2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase inhibitor 1C (Cyclin-dependent kinase inhibitor p57) (p57KIP2). | |||||
|
NU160_MOUSE
|
||||||
| NC score | 0.035837 (rank : 17) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z0W3, Q3TBI7, Q3TP11, Q3U250, Q6A0A7, Q7TME1, Q9CZD9 | Gene names | Nup160, Gtl1-13, Kiaa0197 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin) (Gene trap locus 1-13) (GTL-13). | |||||
|
ELF2_MOUSE
|
||||||
| NC score | 0.030995 (rank : 18) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JHC9, Q6NST2, Q8BTX8, Q9JHC7, Q9JHC8, Q9JHD0 | Gene names | Elf2, Nerf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
AKAP8_MOUSE
|
||||||
| NC score | 0.030783 (rank : 19) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DBR0, Q9R0L8 | Gene names | Akap8, Akap95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 8 (A-kinase anchor protein 95 kDa) (AKAP 95). | |||||
|
NU160_HUMAN
|
||||||
| NC score | 0.030592 (rank : 20) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12769, Q96GB3, Q9H660 | Gene names | NUP160, KIAA0197, NUP120 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear pore complex protein Nup160 (Nucleoporin Nup160) (160 kDa nucleoporin). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.029211 (rank : 21) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.029203 (rank : 22) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.029072 (rank : 23) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.028028 (rank : 24) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.027982 (rank : 25) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CC47_MOUSE
|
||||||
| NC score | 0.024818 (rank : 26) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D024, Q3TSB5, Q3V1S7, Q8C5D9, Q920S6 | Gene names | Ccdc47, Asp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 47 precursor (Adipocyte-specific protein 4). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.023258 (rank : 27) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.023122 (rank : 28) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ELF2_HUMAN
|
||||||
| NC score | 0.023064 (rank : 29) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15723, Q15724, Q15725, Q6P1K5 | Gene names | ELF2, NERF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS-related transcription factor Elf-2 (E74-like factor 2) (New ETS- related factor). | |||||
|
YTHD3_HUMAN
|
||||||
| NC score | 0.020665 (rank : 30) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z739, Q63Z37, Q659A3 | Gene names | YTHDF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
YTHD3_MOUSE
|
||||||
| NC score | 0.020660 (rank : 31) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BYK6, Q3UVI5, Q6NXJ6, Q6NXJ8, Q8BKB6 | Gene names | Ythdf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YTH domain family protein 3. | |||||
|
NUF2_MOUSE
|
||||||
| NC score | 0.019811 (rank : 32) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
K1849_MOUSE
|
||||||
| NC score | 0.017215 (rank : 33) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q69Z89, Q8BNL0, Q8C549, Q8CAL1 | Gene names | Kiaa1849 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1849. | |||||
|
T240L_HUMAN
|
||||||
| NC score | 0.016919 (rank : 34) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71F56, Q68DN4, Q9H8C0, Q9NSY9, Q9UFD8, Q9UPX5 | Gene names | THRAP2, KIAA1025, PROSIT240, TRAP240L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
G3BP2_MOUSE
|
||||||
| NC score | 0.016789 (rank : 35) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97379, Q9R1B8 | Gene names | G3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 2 (GAP SH3-domain- binding protein 2) (G3BP-2). | |||||
|
TTF2_HUMAN
|
||||||
| NC score | 0.016168 (rank : 36) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNY4, O75921, Q5T2K7, Q5VVU8, Q8N6I8 | Gene names | TTF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription termination factor 2 (EC 3.6.1.-) (RNA polymerase II termination factor) (Transcription release factor 2) (Factor 2) (F2) (HuF2) (Lodestar homolog). | |||||
|
TNR6_MOUSE
|
||||||
| NC score | 0.015205 (rank : 37) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25446, Q9DCQ1 | Gene names | Fas, Apt1, Tnfrsf6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.015042 (rank : 38) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.014669 (rank : 39) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.013951 (rank : 40) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.013827 (rank : 41) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.013500 (rank : 42) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
DHX8_HUMAN
|
||||||
| NC score | 0.012673 (rank : 43) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14562 | Gene names | DHX8, DDX8 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DHX8 (EC 3.6.1.-) (DEAH box protein 8) (RNA helicase HRH1). | |||||
|
MYT1L_MOUSE
|
||||||
| NC score | 0.011321 (rank : 44) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97500, O08996, Q8C643, Q8C7L4, Q8CHB4 | Gene names | Myt1l, Kiaa1106, Nzf1, Png1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (Postmitotic neural gene 1 protein) (Zinc finger protein Png-1) (Neural zinc finger factor 1) (NZF-1). | |||||
|
BMP2K_MOUSE
|
||||||
| NC score | 0.010057 (rank : 45) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 874 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Z96, Q8C8L7 | Gene names | Bmp2k, Bike | |||
|
Domain Architecture |

|
|||||
| Description | BMP-2-inducible protein kinase (EC 2.7.11.1) (BIKe). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.009713 (rank : 46) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
ST1C1_MOUSE
|
||||||
| NC score | 0.009608 (rank : 47) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80VR3, O70262 | Gene names | Sult1c1, Sult1a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sulfotransferase 1C1 (EC 2.8.2.-) (Phenol sulfotransferase). | |||||
|
E41L2_MOUSE
|
||||||
| NC score | 0.008466 (rank : 48) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O70318 | Gene names | Epb41l2, Epb4.1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 2 (Generally expressed protein 4.1) (4.1G). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.008169 (rank : 49) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
HCN2_MOUSE
|
||||||
| NC score | 0.006712 (rank : 50) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88703, O70506 | Gene names | Hcn2, Bcng2, Hac1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2) (Hyperpolarization-activated cation channel 1) (HAC-1). | |||||
|
TNIK_HUMAN
|
||||||
| NC score | 0.006224 (rank : 51) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1570 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UKE5, O60298, Q8WUY7, Q9UKD8, Q9UKD9, Q9UKE0, Q9UKE1, Q9UKE2, Q9UKE3, Q9UKE4 | Gene names | TNIK, KIAA0551 | |||
|
Domain Architecture |

|
|||||
| Description | TRAF2 and NCK-interacting protein kinase (EC 2.7.11.1). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.005263 (rank : 52) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
SMUF2_HUMAN
|
||||||
| NC score | 0.004764 (rank : 53) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HAU4, Q9H260 | Gene names | SMURF2 | |||
|
Domain Architecture |

|
|||||
| Description | Smad ubiquitination regulatory factor 2 (EC 6.3.2.-) (Ubiquitin-- protein ligase SMURF2) (Smad-specific E3 ubiquitin ligase 2) (hSMURF2). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.003214 (rank : 54) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.003046 (rank : 55) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.002167 (rank : 56) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||