Please be patient as the page loads
|
DPOD1_HUMAN
|
||||||
| SwissProt Accessions | P28340, Q8NER3, Q96H98 | Gene names | POLD1, POLD | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7) (DNA polymerase subunit delta p125). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DPOD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28340, Q8NER3, Q96H98 | Gene names | POLD1, POLD | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7) (DNA polymerase subunit delta p125). | |||||
|
DPOD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.997062 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P52431, O54883 | Gene names | Pold1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7). | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 2.82168e-86 (rank : 3) | NC score | 0.808089 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
DPOLZ_HUMAN
|
||||||
| θ value | 1.71185e-83 (rank : 4) | NC score | 0.803381 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
DPOLA_MOUSE
|
||||||
| θ value | 3.37202e-47 (rank : 5) | NC score | 0.753661 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOLA_HUMAN
|
||||||
| θ value | 2.85459e-46 (rank : 6) | NC score | 0.758701 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09884, Q86UQ7 | Gene names | POLA1, POLA | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOE1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.151735 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVF7, Q9QX50 | Gene names | Pole, Pole1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
DPOE1_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.151195 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
F120C_MOUSE
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.044590 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
GMEB2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.031674 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
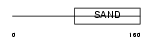
| SwissProt Accessions | Q9UKD1, Q5TDS0, Q9H431, Q9H4X7, Q9H4X8, Q9UF78, Q9ULF1 | Gene names | GMEB2, KIAA1269 | |||
|
Domain Architecture |
|
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2) (Parvovirus initiation factor p79) (PIF p79) (DNA-binding protein p79PIF). | |||||
|
F120C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.043641 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NX05 | Gene names | FAM120C, CXorf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C (Tumor antigen BJ-HCC-21). | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.016718 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.001934 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
COR1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.011029 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
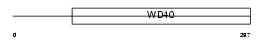
| SwissProt Accessions | Q9WUM3, Q3UEB1, Q9CVA2 | Gene names | Coro1b | |||
|
Domain Architecture |
|
|||||
| Description | Coronin-1B (Coronin-2). | |||||
|
MASP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.005458 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
ZCRB1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.012877 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
K0226_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.017510 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
PDIA4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.022093 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13667 | Gene names | PDIA4, ERP70, ERP72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.011408 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
ISL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.012609 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
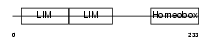
| SwissProt Accessions | P61371, P20663, P47894 | Gene names | ISL1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.012609 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
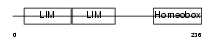
| SwissProt Accessions | P61372, P20663, P47894, Q812D8 | Gene names | Isl1 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
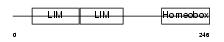
ISL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.012468 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
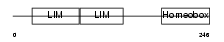
ISL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.012402 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXV0 | Gene names | Isl2 | |||
|
Domain Architecture |
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
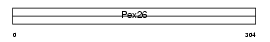
PEX26_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.039693 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGI5, Q3UGH5, Q9D661 | Gene names | Pex26 | |||
|
Domain Architecture |
|
|||||
| Description | Peroxisome assembly protein 26 (Peroxin-26). | |||||
|
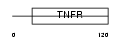
TNR5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.008730 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
CORO6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.007615 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920M5, Q920M3, Q920M4 | Gene names | Coro6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-6 (Clipin E). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.005572 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
PDIA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.015989 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
SGP50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.051770 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q02596 | Gene names | Glycam1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated 50 kDa glycoprotein precursor (SGP50) (Endothelial ligand FOR L-selectin) (Glycosylation-dependent cell adhesion molecule 1) (GlyCAM-1) (MC26). | |||||
|
DPOD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28340, Q8NER3, Q96H98 | Gene names | POLD1, POLD | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7) (DNA polymerase subunit delta p125). | |||||
|
DPOD1_MOUSE
|
||||||
| NC score | 0.997062 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P52431, O54883 | Gene names | Pold1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase delta catalytic subunit (EC 2.7.7.7). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.808089 (rank : 3) | θ value | 2.82168e-86 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
DPOLZ_HUMAN
|
||||||
| NC score | 0.803381 (rank : 4) | θ value | 1.71185e-83 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60673, O43214 | Gene names | REV3L, POLZ, REV3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (hREV3). | |||||
|
DPOLA_HUMAN
|
||||||
| NC score | 0.758701 (rank : 5) | θ value | 2.85459e-46 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09884, Q86UQ7 | Gene names | POLA1, POLA | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOLA_MOUSE
|
||||||
| NC score | 0.753661 (rank : 6) | θ value | 3.37202e-47 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P33609 | Gene names | Pola1, Pola | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase alpha catalytic subunit (EC 2.7.7.7). | |||||
|
DPOE1_MOUSE
|
||||||
| NC score | 0.151735 (rank : 7) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9WVF7, Q9QX50 | Gene names | Pole, Pole1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
DPOE1_HUMAN
|
||||||
| NC score | 0.151195 (rank : 8) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q07864, Q13533, Q86VH9 | Gene names | POLE, POLE1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon, catalytic subunit A (EC 2.7.7.7) (DNA polymerase II subunit A). | |||||
|
SGP50_MOUSE
|
||||||
| NC score | 0.051770 (rank : 9) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q02596 | Gene names | Glycam1 | |||
|
Domain Architecture |

|
|||||
| Description | Sulfated 50 kDa glycoprotein precursor (SGP50) (Endothelial ligand FOR L-selectin) (Glycosylation-dependent cell adhesion molecule 1) (GlyCAM-1) (MC26). | |||||
|
F120C_MOUSE
|
||||||
| NC score | 0.044590 (rank : 10) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C3F2 | Gene names | Fam120c, ORF34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C. | |||||
|
F120C_HUMAN
|
||||||
| NC score | 0.043641 (rank : 11) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NX05 | Gene names | FAM120C, CXorf17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0318 protein FAM120C (Tumor antigen BJ-HCC-21). | |||||
|
PEX26_MOUSE
|
||||||
| NC score | 0.039693 (rank : 12) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGI5, Q3UGH5, Q9D661 | Gene names | Pex26 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisome assembly protein 26 (Peroxin-26). | |||||
|
GMEB2_HUMAN
|
||||||
| NC score | 0.031674 (rank : 13) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UKD1, Q5TDS0, Q9H431, Q9H4X7, Q9H4X8, Q9UF78, Q9ULF1 | Gene names | GMEB2, KIAA1269 | |||
|
Domain Architecture |

|
|||||
| Description | Glucocorticoid modulatory element-binding protein 2 (GMEB-2) (Parvovirus initiation factor p79) (PIF p79) (DNA-binding protein p79PIF). | |||||
|
PDIA4_HUMAN
|
||||||
| NC score | 0.022093 (rank : 14) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13667 | Gene names | PDIA4, ERP70, ERP72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
K0226_MOUSE
|
||||||
| NC score | 0.017510 (rank : 15) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80U62, Q3TAX3, Q3TDF7, Q3TDU2, Q6P9T7, Q6PG18, Q8BMP7, Q8BY22 | Gene names | Kiaa0226 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0226. | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.016718 (rank : 16) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
PDIA4_MOUSE
|
||||||
| NC score | 0.015989 (rank : 17) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08003, P15841 | Gene names | Pdia4, Cai, Erp72 | |||
|
Domain Architecture |

|
|||||
| Description | Protein disulfide-isomerase A4 precursor (EC 5.3.4.1) (Protein ERp-72) (ERp72). | |||||
|
ZCRB1_MOUSE
|
||||||
| NC score | 0.012877 (rank : 18) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CZ96, Q3TDL3 | Gene names | Zcrb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC-type and RNA-binding motif-containing protein 1 (MADP-1). | |||||
|
ISL1_HUMAN
|
||||||
| NC score | 0.012609 (rank : 19) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61371, P20663, P47894 | Gene names | ISL1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL1_MOUSE
|
||||||
| NC score | 0.012609 (rank : 20) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61372, P20663, P47894, Q812D8 | Gene names | Isl1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL2_HUMAN
|
||||||
| NC score | 0.012468 (rank : 21) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
ISL2_MOUSE
|
||||||
| NC score | 0.012402 (rank : 22) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CXV0 | Gene names | Isl2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.011408 (rank : 23) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
COR1B_MOUSE
|
||||||
| NC score | 0.011029 (rank : 24) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WUM3, Q3UEB1, Q9CVA2 | Gene names | Coro1b | |||
|
Domain Architecture |

|
|||||
| Description | Coronin-1B (Coronin-2). | |||||
|
TNR5_MOUSE
|
||||||
| NC score | 0.008730 (rank : 25) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
CORO6_MOUSE
|
||||||
| NC score | 0.007615 (rank : 26) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920M5, Q920M3, Q920M4 | Gene names | Coro6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coronin-6 (Clipin E). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.005572 (rank : 27) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.005458 (rank : 28) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.001934 (rank : 29) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||