Please be patient as the page loads
|
K2027_MOUSE
|
||||||
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K2027_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.905472 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
K2027_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 3) | NC score | 0.487979 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 4.44505e-15 (rank : 4) | NC score | 0.401906 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GRIN2_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 5) | NC score | 0.551388 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60269, Q5SVF0 | Gene names | GPRIN2, KIAA0514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 2 (GRIN2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 6) | NC score | 0.154673 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 7) | NC score | 0.136902 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 8) | NC score | 0.151275 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 9) | NC score | 0.082199 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 10) | NC score | 0.105637 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AKAP6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.080032 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 12) | NC score | 0.027988 (rank : 119) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 13) | NC score | 0.069455 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.089045 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
RTN1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 15) | NC score | 0.066763 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.076403 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
MUC1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.120485 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.083775 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.079808 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.128274 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
RERE_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.064146 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
RHG04_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.029123 (rank : 116) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.102743 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.092306 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.108927 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
UBP42_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.045398 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.047088 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.099588 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.028969 (rank : 117) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.365318 (rank : 30) | NC score | 0.143551 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.067183 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.47712 (rank : 32) | NC score | 0.048351 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 33) | NC score | 0.027435 (rank : 120) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PHF2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 34) | NC score | 0.046772 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.62314 (rank : 35) | NC score | 0.052392 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 36) | NC score | 0.050840 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
41_MOUSE
|
||||||
| θ value | 0.813845 (rank : 37) | NC score | 0.016771 (rank : 135) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 38) | NC score | 0.098779 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.022255 (rank : 125) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
KLF9_MOUSE
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.005326 (rank : 145) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35739 | Gene names | Klf9, Bteb, Bteb-1, Bteb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 9 (Transcription factor BTEB1) (Basic transcription element-binding protein 1) (BTE-binding protein 1) (GC box-binding protein 1). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 41) | NC score | 0.049098 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
EDAR_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.035629 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
GHR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.033459 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10912, Q9HCX2 | Gene names | GHR | |||
|
Domain Architecture |
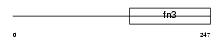
|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.025590 (rank : 122) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NUAK2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.005172 (rank : 146) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
PCX1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.044764 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 47) | NC score | 0.090070 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.022587 (rank : 124) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GGN_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.065137 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.016301 (rank : 137) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.084525 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.069423 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
ALMS1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.036045 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.030486 (rank : 114) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.059244 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HIPK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 56) | NC score | 0.007470 (rank : 144) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
RERE_MOUSE
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.060469 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.036832 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.056373 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HRX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.042151 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
NCOR2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.047832 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.075587 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
SHB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.018241 (rank : 133) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15464, Q504U5, Q5VUM8 | Gene names | SHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein B. | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.068594 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.031580 (rank : 112) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.027324 (rank : 121) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.057301 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CENG1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.018933 (rank : 131) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DIP2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.018349 (rank : 132) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
ELOA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.031515 (rank : 113) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
KLF9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.003889 (rank : 147) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13886, Q16196 | Gene names | KLF9, BTEB, BTEB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 9 (Transcription factor BTEB1) (Basic transcription element-binding protein 1) (BTE-binding protein 1) (GC box-binding protein 1). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.049877 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MYLK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.012074 (rank : 139) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.048650 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.066333 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.036015 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.037711 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.056924 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.040301 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DUS8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 80) | NC score | 0.011837 (rank : 140) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
GLYG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.021099 (rank : 127) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15488, O15485, O15486, O15487, O15489, O15490 | Gene names | GYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycogenin-2 (EC 2.4.1.186) (GN-2) (GN2). | |||||
|
HAX1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.028211 (rank : 118) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35387 | Gene names | Hax1, Hs1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HS1-associating protein X-1 (HAX-1) (HS1-binding protein). | |||||
|
IF4B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.031808 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.063915 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.057602 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
PHF2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.038724 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
STA13_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.016367 (rank : 136) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.023148 (rank : 123) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CN044_HUMAN
|
||||||
| θ value | 6.88961 (rank : 89) | NC score | 0.019240 (rank : 130) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MY7 | Gene names | C14orf44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44. | |||||
|
KU86_MOUSE
|
||||||
| θ value | 6.88961 (rank : 90) | NC score | 0.017857 (rank : 134) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27641 | Gene names | Xrcc5, G22p2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Ku autoantigen protein p86 homolog) (Ku80) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.052240 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.053297 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.058996 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TTC28_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.029148 (rank : 115) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.034702 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANR28_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.002958 (rank : 148) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
EFS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.019882 (rank : 129) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
GR65_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.013907 (rank : 138) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
LIFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.008325 (rank : 143) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42702, Q6LCD9 | Gene names | LIFR | |||
|
Domain Architecture |

|
|||||
| Description | Leukemia inhibitory factor receptor precursor (LIF receptor) (LIF-R) (CD118 antigen). | |||||
|
LMBTL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.009464 (rank : 142) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
M3K7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.000814 (rank : 149) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43318, O43317, O43319, Q5TDN2, Q5TDN3 | Gene names | MAP3K7, TAK1 | |||
|
Domain Architecture |
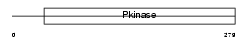
|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.050643 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
PTHR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.049725 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12272, Q15251 | Gene names | PTHLH, PTHRP | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone-related protein precursor (PTH-rP) (PTHrP) [Contains: PTHrP[1-36]; PTHrP[38-94]; Osteostatin (PTHrP[107-139])]. | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.020321 (rank : 128) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
TR240_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.021259 (rank : 126) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
ZN687_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.011375 (rank : 141) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
AFF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.069164 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.066630 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.050912 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.050617 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.075558 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.058990 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.063387 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052212 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
CK024_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051829 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051278 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.054532 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.057609 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.053674 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.051550 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
KI67_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063489 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.065915 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.058010 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052254 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.056212 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.069739 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056617 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.052190 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.079816 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.067409 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MINT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.057303 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.064907 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.094317 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.062515 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.086427 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.061022 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.055862 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051788 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.078445 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.060702 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.059726 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
SELPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.057019 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.071044 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.073401 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.055372 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.051334 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.061662 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.061619 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.051771 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
K2027_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q8BWS5, Q69Z30 | Gene names | Kiaa2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
K2027_HUMAN
|
||||||
| NC score | 0.905472 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q6ZVF9, Q8IVE4 | Gene names | KIAA2027 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA2027. | |||||
|
GRIN2_HUMAN
|
||||||
| NC score | 0.551388 (rank : 3) | θ value | 2.43908e-13 (rank : 5) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60269, Q5SVF0 | Gene names | GPRIN2, KIAA0514 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 2 (GRIN2). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.487979 (rank : 4) | θ value | 3.40345e-15 (rank : 3) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.401906 (rank : 5) | θ value | 4.44505e-15 (rank : 4) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.154673 (rank : 6) | θ value | 0.00298849 (rank : 6) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.151275 (rank : 7) | θ value | 0.00390308 (rank : 8) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.143551 (rank : 8) | θ value | 0.365318 (rank : 30) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.136902 (rank : 9) | θ value | 0.00298849 (rank : 7) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.128274 (rank : 10) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
MUC1_MOUSE
|
||||||
| NC score | 0.120485 (rank : 11) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q02496 | Gene names | Muc1, Muc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (Polymorphic epithelial mucin) (PEMT) (Episialin). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.108927 (rank : 12) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.105637 (rank : 13) | θ value | 0.0193708 (rank : 10) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.102743 (rank : 14) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.099588 (rank : 15) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.098779 (rank : 16) | θ value | 0.813845 (rank : 38) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.094317 (rank : 17) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.092306 (rank : 18) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.090070 (rank : 19) | θ value | 1.38821 (rank : 47) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.089045 (rank : 20) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.086427 (rank : 21) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.084525 (rank : 22) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.083775 (rank : 23) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.082199 (rank : 24) | θ value | 0.00869519 (rank : 9) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
AKAP6_HUMAN
|
||||||
| NC score | 0.080032 (rank : 25) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13023, O15028 | Gene names | AKAP6, AKAP100, KIAA0311 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 6 (Protein kinase A-anchoring protein 6) (PRKA6) (A-kinase anchor protein 100 kDa) (AKAP 100) (mAKAP). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.079816 (rank : 26) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.079808 (rank : 27) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.078445 (rank : 28) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.076403 (rank : 29) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.075587 (rank : 30) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.075558 (rank : 31) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.073401 (rank : 32) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.071044 (rank : 33) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.069739 (rank : 34) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.069455 (rank : 35) | θ value | 0.0431538 (rank : 13) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.069423 (rank : 36) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.069164 (rank : 37) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.068594 (rank : 38) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.067409 (rank : 39) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.067183 (rank : 40) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
RTN1_MOUSE
|
||||||
| NC score | 0.066763 (rank : 41) | θ value | 0.0736092 (rank : 15) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K0T0, Q8K4S4 | Gene names | Rtn1, Nsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reticulon-1 (Neuroendocrine-specific protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.066630 (rank : 42) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.066333 (rank : 43) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.065915 (rank : 44) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.065137 (rank : 45) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.064907 (rank : 46) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RERE_HUMAN
|
||||||
| NC score | 0.064146 (rank : 47) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 779 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9P2R6, O43393, O75046, O75359, Q5VXL9, Q6P6B9, Q9Y2W4 | Gene names | RERE, ARG, ARP, ATN1L, KIAA0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-1-like protein) (Atrophin-1-related protein). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.063915 (rank : 48) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.063489 (rank : 49) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.063387 (rank : 50) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.062515 (rank : 51) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.061662 (rank : 52) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.061619 (rank : 53) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.061022 (rank : 54) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.060702 (rank : 55) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.060469 (rank : 56) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.059726 (rank : 57) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.059244 (rank : 58) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.058996 (rank : 59) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.058990 (rank : 60) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.058010 (rank : 61) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.057609 (rank : 62) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.057602 (rank : 63) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.057303 (rank : 64) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.057301 (rank : 65) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.057019 (rank : 66) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.056924 (rank : 67) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.056617 (rank : 68) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.056373 (rank : 69) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.056212 (rank : 70) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.055862 (rank : 71) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.055372 (rank : 72) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.054532 (rank : 73) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.053674 (rank : 74) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.053297 (rank : 75) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.052392 (rank : 76) | θ value | 0.62314 (rank : 35) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.052254 (rank : 77) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.052240 (rank : 78) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.052212 (rank : 79) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.052190 (rank : 80) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
CK024_MOUSE
|
||||||
| NC score | 0.051829 (rank : 81) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9D8N1, Q8VCP2 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 homolog precursor. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.051788 (rank : 82) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.051771 (rank : 83) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.051550 (rank : 84) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.051334 (rank : 85) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.051278 (rank : 86) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.050912 (rank : 87) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.050840 (rank : 88) | θ value | 0.62314 (rank : 36) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.050643 (rank : 89) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.050617 (rank : 90) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.049877 (rank : 91) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PTHR_HUMAN
|
||||||
| NC score | 0.049725 (rank : 92) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12272, Q15251 | Gene names | PTHLH, PTHRP | |||
|
Domain Architecture |

|
|||||
| Description | Parathyroid hormone-related protein precursor (PTH-rP) (PTHrP) [Contains: PTHrP[1-36]; PTHrP[38-94]; Osteostatin (PTHrP[107-139])]. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.049098 (rank : 93) | θ value | 1.06291 (rank : 41) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.048650 (rank : 94) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.048351 (rank : 95) | θ value | 0.47712 (rank : 32) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NCOR2_HUMAN
|
||||||
| NC score | 0.047832 (rank : 96) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 870 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y618, O00613, O15416, Q13354, Q9Y5U0 | Gene names | NCOR2, CTG26 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 2 (N-CoR2) (Silencing mediator of retinoic acid and thyroid hormone receptor) (SMRT) (SMRTe) (Thyroid-, retinoic-acid-receptor-associated corepressor) (T3 receptor- associating factor) (TRAC) (CTG repeat protein 26) (SMAP270). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.047088 (rank : 97) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
PHF2_MOUSE
|
||||||
| NC score | 0.046772 (rank : 98) | θ value | 0.47712 (rank : 34) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9WTU0, Q80WA8 | Gene names | Phf2 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
UBP42_HUMAN
|
||||||
| NC score | 0.045398 (rank : 99) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9H9J4, Q3C166, Q6P9B4 | Gene names | USP42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 42 (EC 3.1.2.15) (Ubiquitin thioesterase 42) (Ubiquitin-specific-processing protease 42) (Deubiquitinating enzyme 42). | |||||
|
PCX1_HUMAN
|
||||||
| NC score | 0.044764 (rank : 100) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96RV3, O94897, Q96AI7, Q9Y2J9 | Gene names | PCNX, KIAA0805, KIAA0995, PCNXL1 | |||
|
Domain Architecture |

|
|||||
| Description | Pecanex-like protein 1 (Pecanex homolog). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.042151 (rank : 101) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.040301 (rank : 102) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
PHF2_HUMAN
|
||||||
| NC score | 0.038724 (rank : 103) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O75151, Q8N3K2, Q9Y6N4 | Gene names | PHF2, KIAA0662 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 2 (GRC5). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.037711 (rank : 104) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.036832 (rank : 105) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
ALMS1_MOUSE
|
||||||
| NC score | 0.036045 (rank : 106) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K4E0, Q6A084, Q8C9N9 | Gene names | Alms1, Kiaa0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1 homolog. | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.036015 (rank : 107) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
EDAR_MOUSE
|
||||||
| NC score | 0.035629 (rank : 108) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.034702 (rank : 109) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
GHR_HUMAN
|
||||||
| NC score | 0.033459 (rank : 110) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10912, Q9HCX2 | Gene names | GHR | |||
|
Domain Architecture |

|
|||||
| Description | Growth hormone receptor precursor (GH receptor) (Somatotropin receptor) [Contains: Growth hormone-binding protein (GH-binding protein) (GHBP) (Serum-binding protein)]. | |||||
|
IF4B_HUMAN
|
||||||
| NC score | 0.031808 (rank : 111) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P23588 | Gene names | EIF4B | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4B (eIF-4B). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.031580 (rank : 112) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ELOA3_HUMAN
|
||||||
| NC score | 0.031515 (rank : 113) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NG57 | Gene names | TCEB3C, TCEB3L2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II transcription factor SIII subunit A3 (Elongin-A3) (EloA3) (Transcription elongation factor B polypeptide 3C). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.030486 (rank : 114) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
TTC28_MOUSE
|
||||||
| NC score | 0.029148 (rank : 115) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80XJ3, Q6P9J6, Q80TL5, Q8BV10, Q8C0F2 | Gene names | Ttc28, Kiaa1043 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 28 (TPR repeat protein 28). | |||||
|
RHG04_HUMAN
|
||||||
| NC score | 0.029123 (rank : 116) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P98171, Q14144 | Gene names | ARHGAP4, KIAA0131, RGC1, RHOGAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 4 (Rho-GAP hematopoietic protein C1) (p115). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.028969 (rank : 117) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
HAX1_MOUSE
|
||||||
| NC score | 0.028211 (rank : 118) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35387 | Gene names | Hax1, Hs1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HS1-associating protein X-1 (HAX-1) (HS1-binding protein). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.027988 (rank : 119) | θ value | 0.0431538 (rank : 12) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.027435 (rank : 120) | θ value | 0.47712 (rank : 33) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.027324 (rank : 121) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.025590 (rank : 122) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.023148 (rank : 123) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.022587 (rank : 124) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.022255 (rank : 125) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
TR240_HUMAN
|
||||||
| NC score | 0.021259 (rank : 126) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UHV7, O60334 | Gene names | THRAP1, ARC250, KIAA0593, TRAP240 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid hormone receptor-associated protein complex 240 kDa component (Trap240) (Thyroid hormone receptor-associated protein 1) (Vitamin D3 receptor-interacting protein complex component DRIP250) (DRIP 250) (Activator-recruited cofactor 250 kDa component) (ARC250). | |||||
|
GLYG2_HUMAN
|
||||||
| NC score | 0.021099 (rank : 127) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15488, O15485, O15486, O15487, O15489, O15490 | Gene names | GYG2 | |||
|
Domain Architecture |

|
|||||
| Description | Glycogenin-2 (EC 2.4.1.186) (GN-2) (GN2). | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.020321 (rank : 128) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
EFS_HUMAN
|
||||||
| NC score | 0.019882 (rank : 129) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O43281, O43282 | Gene names | EFS | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (HEFS). | |||||
|
CN044_HUMAN
|
||||||
| NC score | 0.019240 (rank : 130) | θ value | 6.88961 (rank : 89) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MY7 | Gene names | C14orf44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf44. | |||||
|
CENG1_MOUSE
|
||||||
| NC score | 0.018933 (rank : 131) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3UHD9, Q5DU45 | Gene names | Centg1, Agap2, Kiaa0167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centaurin-gamma 1 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 2) (AGAP-2) (Phosphatidylinositol-3-kinase enhancer) (PIKE). | |||||
|
DIP2A_HUMAN
|
||||||
| NC score | 0.018349 (rank : 132) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14689, Q8IVA3, Q8N4S2, Q8TD89, Q96ML9 | Gene names | DIP2A, C21orf106, DIP2, KIAA0184 | |||
|
Domain Architecture |

|
|||||
| Description | Disco-interacting protein 2 homolog A. | |||||
|
SHB_HUMAN
|
||||||
| NC score | 0.018241 (rank : 133) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15464, Q504U5, Q5VUM8 | Gene names | SHB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing adapter protein B. | |||||
|
KU86_MOUSE
|
||||||
| NC score | 0.017857 (rank : 134) | θ value | 6.88961 (rank : 90) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27641 | Gene names | Xrcc5, G22p2 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase 2 subunit 2 (EC 3.6.1.-) (ATP-dependent DNA helicase II 80 kDa subunit) (Ku autoantigen protein p86 homolog) (Ku80) (CTC box-binding factor 85 kDa subunit) (CTCBF) (CTC85) (Nuclear factor IV) (DNA-repair protein XRCC5). | |||||
|
41_MOUSE
|
||||||
| NC score | 0.016771 (rank : 135) | θ value | 0.813845 (rank : 37) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48193, Q68FF1, Q6NVF5 | Gene names | Epb41, Epb4.1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (4.1R). | |||||
|
STA13_HUMAN
|
||||||
| NC score | 0.016367 (rank : 136) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3M8, Q5HYH1, Q5TAE3, Q6UN61, Q86TP6, Q86WQ3, Q86XT1 | Gene names | STARD13, DLC2, GT650 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 13 (StARD13) (START domain- containing protein 13) (46H23.2) (Deleted in liver cancer protein 2) (Rho GTPase-activating protein). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.016301 (rank : 137) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.013907 (rank : 138) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
MYLK_MOUSE
|
||||||
| NC score | 0.012074 (rank : 139) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1489 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PDN3, Q3TSJ7, Q80UX0, Q80YN7, Q80YN8, Q8K026, Q924D2, Q9ERD3 | Gene names | Mylk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin light chain kinase, smooth muscle (EC 2.7.11.18) (MLCK) (Telokin) (Kinase-related protein) (KRP). | |||||
|
DUS8_HUMAN
|
||||||
| NC score | 0.011837 (rank : 140) | θ value | 5.27518 (rank : 80) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13202 | Gene names | DUSP8, VH5 | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity protein phosphatase 8 (EC 3.1.3.48) (EC 3.1.3.16) (Dual specificity protein phosphatase hVH-5). | |||||
|
ZN687_HUMAN
|
||||||
| NC score | 0.011375 (rank : 141) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 1244 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N1G0, Q68DQ8, Q9H937, Q9P2A7 | Gene names | ZNF687, KIAA1441 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
LMBTL_HUMAN
|
||||||
| NC score | 0.009464 (rank : 142) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y468, Q5H8Y8, Q8IUV7, Q9H1E6, Q9H1G5, Q9UG06, Q9UJB9, Q9Y4C9 | Gene names | L3MBTL, KIAA0681, L3MBT | |||
|
Domain Architecture |

|
|||||
| Description | Lethal(3)malignant brain tumor-like protein (L(3)mbt-like) (L(3)mbt protein homolog) (H-l(3)mbt protein) (H-L(3)MBT) (L3MBTL1). | |||||
|
LIFR_HUMAN
|
||||||
| NC score | 0.008325 (rank : 143) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P42702, Q6LCD9 | Gene names | LIFR | |||
|
Domain Architecture |

|
|||||
| Description | Leukemia inhibitory factor receptor precursor (LIF receptor) (LIF-R) (CD118 antigen). | |||||
|
HIPK1_HUMAN
|
||||||
| NC score | 0.007470 (rank : 144) | θ value | 2.36792 (rank : 56) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Z02, O75125, Q8IYD7, Q8NDN5, Q8NEB6, Q8TBZ1 | Gene names | HIPK1, KIAA0630 | |||
|
Domain Architecture |

|
|||||
| Description | Homeodomain-interacting protein kinase 1 (EC 2.7.11.1). | |||||
|
KLF9_MOUSE
|
||||||
| NC score | 0.005326 (rank : 145) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35739 | Gene names | Klf9, Bteb, Bteb-1, Bteb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 9 (Transcription factor BTEB1) (Basic transcription element-binding protein 1) (BTE-binding protein 1) (GC box-binding protein 1). | |||||
|
NUAK2_MOUSE
|
||||||
| NC score | 0.005172 (rank : 146) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 878 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BZN4, Q80ZW3, Q8CIC0, Q9DBV0 | Gene names | Nuak2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NUAK family SNF1-like kinase 2 (EC 2.7.11.1). | |||||
|
KLF9_HUMAN
|
||||||
| NC score | 0.003889 (rank : 147) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 697 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13886, Q16196 | Gene names | KLF9, BTEB, BTEB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 9 (Transcription factor BTEB1) (Basic transcription element-binding protein 1) (BTE-binding protein 1) (GC box-binding protein 1). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.002958 (rank : 148) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
M3K7_HUMAN
|
||||||
| NC score | 0.000814 (rank : 149) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 106 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43318, O43317, O43319, Q5TDN2, Q5TDN3 | Gene names | MAP3K7, TAK1 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7 (EC 2.7.11.25) (Transforming growth factor-beta-activated kinase 1) (TGF-beta- activated kinase 1). | |||||