Please be patient as the page loads
|
CBX6_MOUSE
|
||||||
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CBX6_MOUSE
|
||||||
| θ value | 1.23469e-174 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_HUMAN
|
||||||
| θ value | 4.12933e-154 (rank : 2) | NC score | 0.976218 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX8_HUMAN
|
||||||
| θ value | 6.17384e-33 (rank : 3) | NC score | 0.891943 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX8_MOUSE
|
||||||
| θ value | 6.17384e-33 (rank : 4) | NC score | 0.895731 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX4_HUMAN
|
||||||
| θ value | 2.42779e-29 (rank : 5) | NC score | 0.853856 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| θ value | 1.02001e-27 (rank : 6) | NC score | 0.849880 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX7_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 7) | NC score | 0.901308 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 8) | NC score | 0.902589 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX2_HUMAN
|
||||||
| θ value | 5.42112e-21 (rank : 9) | NC score | 0.759742 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 10) | NC score | 0.807575 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 11) | NC score | 0.621407 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 12) | NC score | 0.620349 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 13) | NC score | 0.622038 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | 4.45536e-07 (rank : 14) | NC score | 0.622038 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 15) | NC score | 0.396176 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 16) | NC score | 0.591615 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 17) | NC score | 0.578818 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 0.000121331 (rank : 18) | NC score | 0.386377 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 19) | NC score | 0.369995 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 20) | NC score | 0.370803 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 21) | NC score | 0.212345 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 22) | NC score | 0.292379 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 23) | NC score | 0.384919 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 24) | NC score | 0.310013 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 0.000602161 (rank : 25) | NC score | 0.323269 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 26) | NC score | 0.160603 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
CHD1_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 27) | NC score | 0.167536 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 28) | NC score | 0.165950 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 29) | NC score | 0.299209 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
PRRX2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.012655 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06348, Q61651 | Gene names | Prrx2, Prx2, S8 | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2 (PRX-2) (Homeobox protein S8). | |||||
|
GRIN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.018016 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.014512 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.079030 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CMTA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.018215 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
CAN10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.010059 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | -0.000352 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
TENR_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.004811 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
2DMB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.004559 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35737 | Gene names | H2-DMb1, H-2Mb1, Mb | |||
|
Domain Architecture |
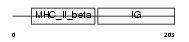
|
|||||
| Description | Class II histocompatibility antigen, M beta 1 chain precursor (H2-M beta 1 chain). | |||||
|
CHD9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.058734 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.058388 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.008047 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
CCD91_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.005762 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
RFX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.007892 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.046885 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.013370 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.066337 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.066084 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
JIP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.007504 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.009783 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
TCF19_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.020654 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
AUHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.056571 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.055917 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.061605 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.057747 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.053842 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
ECH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.071138 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.073279 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.069138 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.069545 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
MS3L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.073617 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
PECI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.106311 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.105710 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CBX6_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.23469e-174 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9DBY5, Q8BZH5 | Gene names | Cbx6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX6_HUMAN
|
||||||
| NC score | 0.976218 (rank : 2) | θ value | 4.12933e-154 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O95503, Q96EM5 | Gene names | CBX6 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 6. | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.902589 (rank : 3) | θ value | 8.36355e-22 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX7_HUMAN
|
||||||
| NC score | 0.901308 (rank : 4) | θ value | 2.96777e-27 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95931, Q86T17 | Gene names | CBX7 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
CBX8_MOUSE
|
||||||
| NC score | 0.895731 (rank : 5) | θ value | 6.17384e-33 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QXV1 | Gene names | Cbx8, Pc3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (mPc3). | |||||
|
CBX8_HUMAN
|
||||||
| NC score | 0.891943 (rank : 6) | θ value | 6.17384e-33 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9HC52, Q96H39, Q9NR07 | Gene names | CBX8, PC3, RC1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 8 (Polycomb 3 homolog) (Pc3) (hPc3) (Rectachrome 1). | |||||
|
CBX4_HUMAN
|
||||||
| NC score | 0.853856 (rank : 7) | θ value | 2.42779e-29 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O00257, Q96C04 | Gene names | CBX4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (hPc2). | |||||
|
CBX4_MOUSE
|
||||||
| NC score | 0.849880 (rank : 8) | θ value | 1.02001e-27 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O55187 | Gene names | Cbx4, Pc2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 4 (Polycomb 2 homolog) (Pc2) (mPc2). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.807575 (rank : 9) | θ value | 5.42112e-21 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CBX2_HUMAN
|
||||||
| NC score | 0.759742 (rank : 10) | θ value | 5.42112e-21 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14781, Q9BTB1 | Gene names | CBX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromobox protein homolog 2. | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.622038 (rank : 11) | θ value | 4.45536e-07 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.622038 (rank : 12) | θ value | 4.45536e-07 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.621407 (rank : 13) | θ value | 3.08544e-08 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.620349 (rank : 14) | θ value | 4.0297e-08 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.591615 (rank : 15) | θ value | 2.44474e-05 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.578818 (rank : 16) | θ value | 0.000121331 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.396176 (rank : 17) | θ value | 6.43352e-06 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.386377 (rank : 18) | θ value | 0.000121331 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.384919 (rank : 19) | θ value | 0.00035302 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.370803 (rank : 20) | θ value | 0.00020696 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.369995 (rank : 21) | θ value | 0.00020696 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.323269 (rank : 22) | θ value | 0.000602161 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.310013 (rank : 23) | θ value | 0.000461057 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.299209 (rank : 24) | θ value | 0.0148317 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.292379 (rank : 25) | θ value | 0.00020696 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.212345 (rank : 26) | θ value | 0.00020696 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
CHD1_HUMAN
|
||||||
| NC score | 0.167536 (rank : 27) | θ value | 0.00175202 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O14646 | Gene names | CHD1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD1_MOUSE
|
||||||
| NC score | 0.165950 (rank : 28) | θ value | 0.00175202 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 266 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P40201 | Gene names | Chd1, Chd-1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 1 (EC 3.6.1.-) (ATP- dependent helicase CHD1) (CHD-1). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.160603 (rank : 29) | θ value | 0.00102713 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.106311 (rank : 30) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.105710 (rank : 31) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.079030 (rank : 32) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
MS3L1_MOUSE
|
||||||
| NC score | 0.073617 (rank : 33) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVG9 | Gene names | Msl3l1, Msl31 | |||
|
Domain Architecture |

|
|||||
| Description | Male-specific lethal 3-like 1 (MSL3-like 1) (Male-specific lethal-3 homolog 1). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.073279 (rank : 34) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.071138 (rank : 35) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.069545 (rank : 36) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.069138 (rank : 37) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.066337 (rank : 38) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.066084 (rank : 39) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.061605 (rank : 40) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD9_HUMAN
|
||||||
| NC score | 0.058734 (rank : 41) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3L8U1, O15025, Q1WF12, Q6DTK9, Q9H9V7 | Gene names | CHD9, KIAA0308, KISH2, PRIC320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Chromatin-related mesenchymal modulator) (CReMM) (Chromatin remodeling factor CHROM1) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha-interacting complex protein 320 kDa) (Kismet homolog 2). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.058388 (rank : 42) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.057747 (rank : 43) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.056571 (rank : 44) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.055917 (rank : 45) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.053842 (rank : 46) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.046885 (rank : 47) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
TCF19_HUMAN
|
||||||
| NC score | 0.020654 (rank : 48) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y242, Q13176, Q15967, Q5SQ89, Q5STD6, Q5STF5, Q9BUM2, Q9UBH7 | Gene names | TCF19, SC1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 19 (Transcription factor SC1). | |||||
|
CMTA1_HUMAN
|
||||||
| NC score | 0.018215 (rank : 49) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6Y1, Q5VUE1 | Gene names | CAMTA1, KIAA0833 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calmodulin-binding transcription activator 1. | |||||
|
GRIN1_HUMAN
|
||||||
| NC score | 0.018016 (rank : 50) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7Z2K8, Q8ND74, Q96PZ4 | Gene names | GPRIN1, KIAA1893 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.014512 (rank : 51) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.013370 (rank : 52) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PRRX2_MOUSE
|
||||||
| NC score | 0.012655 (rank : 53) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06348, Q61651 | Gene names | Prrx2, Prx2, S8 | |||
|
Domain Architecture |

|
|||||
| Description | Paired mesoderm homeobox protein 2 (PRX-2) (Homeobox protein S8). | |||||
|
CAN10_HUMAN
|
||||||
| NC score | 0.010059 (rank : 54) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HC96, Q8NCD4, Q96IG4, Q96JI2, Q9HC89, Q9HC90, Q9HC91, Q9HC92, Q9HC93, Q9HC94, Q9HC95 | Gene names | CAPN10, KIAA1845 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-10 (EC 3.4.22.-) (Calcium-activated neutral proteinase 10) (CANP 10). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.009783 (rank : 55) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.008047 (rank : 56) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
RFX1_HUMAN
|
||||||
| NC score | 0.007892 (rank : 57) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P22670 | Gene names | RFX1 | |||
|
Domain Architecture |

|
|||||
| Description | MHC class II regulatory factor RFX1 (RFX) (Enhancer factor C) (EF-C). | |||||
|
JIP1_HUMAN
|
||||||
| NC score | 0.007504 (rank : 58) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UQF2, O43407 | Gene names | MAPK8IP1, IB1, JIP1 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 1 (JNK-interacting protein 1) (JIP-1) (JNK MAP kinase scaffold protein 1) (Islet-brain 1) (IB-1) (Mitogen-activated protein kinase 8-interacting protein 1). | |||||
|
CCD91_MOUSE
|
||||||
| NC score | 0.005762 (rank : 59) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 699 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D8L5, Q8R3N8, Q9D9E8 | Gene names | Ccdc91, Ggabp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner). | |||||
|
TENR_MOUSE
|
||||||
| NC score | 0.004811 (rank : 60) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BYI9, O88717 | Gene names | Tnr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tenascin-R precursor (TN-R) (Restrictin) (Janusin) (Neural recognition molecule J1-160/180). | |||||
|
2DMB_MOUSE
|
||||||
| NC score | 0.004559 (rank : 61) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35737 | Gene names | H2-DMb1, H-2Mb1, Mb | |||
|
Domain Architecture |

|
|||||
| Description | Class II histocompatibility antigen, M beta 1 chain precursor (H2-M beta 1 chain). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | -0.000352 (rank : 62) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||