Please be patient as the page loads
|
ZN185_HUMAN
|
||||||
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ZN185_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
ZN185_MOUSE
|
||||||
| θ value | 1.07473e-77 (rank : 2) | NC score | 0.894964 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62394 | Gene names | Znf185, Zfp185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein Zfp185) (P1-A). | |||||
|
SCEL_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 3) | NC score | 0.499291 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
SCEL_HUMAN
|
||||||
| θ value | 4.59992e-12 (rank : 4) | NC score | 0.492551 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95171 | Gene names | SCEL | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 3.77169e-06 (rank : 5) | NC score | 0.261445 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
LMO3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 6) | NC score | 0.176388 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAP4, Q8N974, Q9UDD5 | Gene names | LMO3, RBTN3, RHOM3 | |||
|
Domain Architecture |
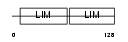
|
|||||
| Description | LIM-only protein 3 (Neuronal-specific transcription factor DAT1) (Rhombotin-3). | |||||
|
LMO3_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 7) | NC score | 0.176388 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BZL8 | Gene names | Lmo3 | |||
|
Domain Architecture |
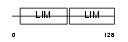
|
|||||
| Description | LIM-only protein 3 (Neuronal-specific transcription factor DAT1). | |||||
|
RBTN1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.177587 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P25800, Q4VBC5 | Gene names | LMO1, RBTN1, RHOM1, TTG1 | |||
|
Domain Architecture |
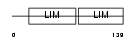
|
|||||
| Description | Rhombotin-1 (Cysteine-rich protein TTG-1) (T-cell translocation protein 1) (LIM-only protein 1). | |||||
|
RBTN1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.177475 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q924W9 | Gene names | Lmo1, Rbtn1, Rhom1, Ttg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhombotin-1 (Cysteine-rich protein TTG-1) (LIM-only protein 1). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 10) | NC score | 0.045293 (rank : 127) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LMO6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 11) | NC score | 0.199222 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80VL3, Q8CAY9 | Gene names | Lmo6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
PRIC2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.205991 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.067458 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
FHL2_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.174423 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O70433, P97448 | Gene names | Fhl2 | |||
|
Domain Architecture |
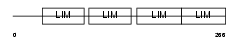
|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3). | |||||
|
LMO6_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.197934 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |
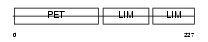
|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 16) | NC score | 0.078797 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
FHL2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 17) | NC score | 0.179876 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14192, Q13229, Q13644, Q9P294 | Gene names | FHL2, DRAL, SLIM3 | |||
|
Domain Architecture |
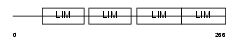
|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3) (LIM-domain protein DRAL). | |||||
|
PRIC2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.202573 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
CLOCK_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 19) | NC score | 0.065451 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
CSRP2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.199934 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16527, Q93030 | Gene names | CSRP2, LMO5, SMLIM | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Smooth muscle cell LIM protein) (SmLIM) (LIM-only protein 5). | |||||
|
CSRP2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.200031 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97314, Q9CZG5 | Gene names | Csrp2, Dlp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.029578 (rank : 145) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
FHL5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.190258 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTX7 | Gene names | Fhl5, Act | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
PDLI5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 24) | NC score | 0.174219 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
CJ092_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.086936 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
CSRP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.200179 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 27) | NC score | 0.200963 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
FHL5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 28) | NC score | 0.179055 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5TD97, Q5TD98, Q8WW21, Q9NQU2 | Gene names | FHL5, ACT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 29) | NC score | 0.176279 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 30) | NC score | 0.061538 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.103048 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.032245 (rank : 143) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.031878 (rank : 144) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
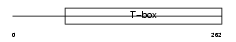
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
BBX_MOUSE
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.044444 (rank : 128) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
CSRP3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.190214 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50462 | Gene names | Csrp3, Clp, Mlp | |||
|
Domain Architecture |
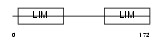
|
|||||
| Description | Cysteine and glycine-rich protein 3 (Cysteine-rich protein 3) (CRP3) (LIM domain protein, cardiac) (Muscle LIM protein). | |||||
|
LDB3_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.174739 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
PRIC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 37) | NC score | 0.193998 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MT3, Q71QF8, Q96N00 | Gene names | PRICKLE1, RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 1 (REST/NRSF-interacting LIM domain protein 1). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.175159 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.041031 (rank : 135) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ZN447_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.020306 (rank : 156) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
CSRP3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.188382 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50461, Q9P131 | Gene names | CSRP3, CLP, MLP | |||
|
Domain Architecture |
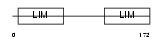
|
|||||
| Description | Cysteine and glycine-rich protein 3 (Cysteine-rich protein 3) (CRP3) (LIM domain protein, cardiac) (Muscle LIM protein). | |||||
|
FHL3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.189895 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R059, Q9JLP5, Q9WUH3 | Gene names | Fhl3 | |||
|
Domain Architecture |
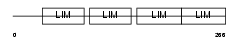
|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
SF3B1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.084282 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
LIPB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.024056 (rank : 153) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.082568 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 46) | NC score | 0.044215 (rank : 129) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FHL3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 47) | NC score | 0.197139 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13643 | Gene names | FHL3, SLIM2 | |||
|
Domain Architecture |
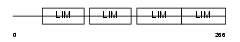
|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 1.06291 (rank : 48) | NC score | 0.047318 (rank : 126) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
PDLI7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.171176 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SC10A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.014716 (rank : 165) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.053789 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
TAU_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.055742 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ZN488_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.039515 (rank : 138) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96MN9 | Gene names | ZNF488 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 488. | |||||
|
ZN553_MOUSE
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.009303 (rank : 171) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3US17, Q3UFP3, Q8R0V0, Q921H7 | Gene names | Znf553, Zfp553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 553. | |||||
|
FBLI1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.170889 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |
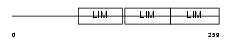
|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.050118 (rank : 123) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
SF3B1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.080640 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.056821 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
FHL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.174540 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97447, O55181, Q8K318 | Gene names | Fhl1 | |||
|
Domain Architecture |
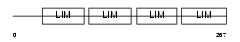
|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM) (KyoT) (RBP-associated molecule 14-1) (RAM14-1). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.168184 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.043749 (rank : 130) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
ZN553_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.009200 (rank : 172) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96MX3, Q4G0R3, Q69YP3, Q96IL9 | Gene names | ZNF553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 553. | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.051578 (rank : 121) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
LMO4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.149226 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61968, O00158, O88894 | Gene names | LMO4 | |||
|
Domain Architecture |
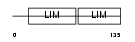
|
|||||
| Description | LIM domain transcription factor LMO4 (LIM domain only protein 4) (LMO- 4) (Breast tumor autoantigen). | |||||
|
LMO4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 65) | NC score | 0.149226 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61969, O00158, O88894 | Gene names | Lmo4 | |||
|
Domain Architecture |
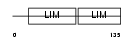
|
|||||
| Description | LIM domain transcription factor LMO4 (LIM domain only protein 4) (LMO- 4) (Breast tumor autoantigen). | |||||
|
TBX15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 66) | NC score | 0.026484 (rank : 147) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TM108_HUMAN
|
||||||
| θ value | 2.36792 (rank : 67) | NC score | 0.034712 (rank : 140) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.035702 (rank : 139) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.043100 (rank : 133) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.015978 (rank : 161) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.026181 (rank : 149) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
ISK6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.040545 (rank : 136) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
S4A7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.014919 (rank : 163) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6M7, O60350, Q6AHZ9, Q9HC88, Q9UIB9 | Gene names | SLC4A7, BT, NBC2, NBC2B, NBC3, SBC2, SLC4A6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium bicarbonate cotransporter 3 (Sodium bicarbonate cotransporter 2) (Sodium bicarbonate cotransporter 2b) (Bicarbonate transporter) (Solute carrier family 4 member 7). | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.007929 (rank : 173) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ZN271_HUMAN
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.007831 (rank : 174) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14591, Q96T29, Q9BSX2, Q9UN33, Q9Y5B7 | Gene names | ZNF271, ZNFEB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 271 (Zinc finger protein HZF7) (Zinc finger protein ZNFphex133) (Epstein-Barr virus-induced zinc finger protein) (ZNF-EB) (CT-ZFP48) (Zinc finger protein dp) (ZNF-dp). | |||||
|
ABLM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.133582 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
CG1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.041361 (rank : 134) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
MAP4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.048576 (rank : 124) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
NGN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.015126 (rank : 162) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.014789 (rank : 164) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 81) | NC score | 0.050608 (rank : 122) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
UBP34_HUMAN
|
||||||
| θ value | 4.03905 (rank : 82) | NC score | 0.014403 (rank : 167) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
BCL6B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.005580 (rank : 176) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N143, Q6PCB4 | Gene names | BCL6B, BAZF, ZNF62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 6 member B protein (Bcl6-associated zinc finger protein) (Zinc finger protein 62). | |||||
|
CDT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.025654 (rank : 151) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
CF049_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.169337 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q2TBC4, Q5T3D4, Q9NSV1 | Gene names | C6orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf49. | |||||
|
CTR9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.025671 (rank : 150) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.026340 (rank : 148) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LHX6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.104618 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88706 | Gene names | Lhx6 | |||
|
Domain Architecture |
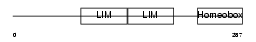
|
|||||
| Description | LIM/homeobox protein Lhx6. | |||||
|
SOX30_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.022291 (rank : 154) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.060854 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.027128 (rank : 146) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ABLM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.134003 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.033187 (rank : 141) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
CBX2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.016038 (rank : 160) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CHSS3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.014578 (rank : 166) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.012383 (rank : 168) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
LAX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.024584 (rank : 152) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHB3, Q8BS18, Q8CGV1 | Gene names | Lax1, Lax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte transmembrane adapter 1 (Membrane-associated adapter protein LAX) (Linker for activation of X cells). | |||||
|
LHX8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.107047 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35652, O70163, O88707 | Gene names | Lhx8, Lhx7 | |||
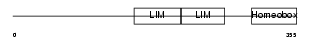
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx8 (LIM homeodomain Lhx7) (L3). | |||||
|
LIMK1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.040291 (rank : 137) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53668 | Gene names | Limk1, Limk | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 1 (EC 2.7.11.1) (LIMK-1) (KIZ-1). | |||||
|
LPXN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.161108 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60711 | Gene names | LPXN, LDLP | |||
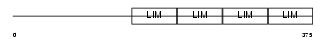
|
Domain Architecture |
|
|||||
| Description | Leupaxin. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.043449 (rank : 131) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 102) | NC score | 0.048116 (rank : 125) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 103) | NC score | 0.016190 (rank : 159) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SALL2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 104) | NC score | 0.011001 (rank : 170) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
CO039_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.021638 (rank : 155) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.032483 (rank : 142) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.000434 (rank : 177) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.012100 (rank : 169) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SOX13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.016211 (rank : 158) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.043428 (rank : 132) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TSSC4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.019024 (rank : 157) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5U2, Q86VL2, Q9BRS6 | Gene names | TSSC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4 (Tumor-suppressing subchromosomal transferable fragment candidate gene 4 protein) (Tumor-suppressing STF cDNA 4 protein). | |||||
|
ZN181_HUMAN
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.005767 (rank : 175) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q2M3W8, Q49A75 | Gene names | ZNF181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 181 (HHZ181). | |||||
|
ABLM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.130875 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6H8Q1, Q6H8Q0, Q6NX73, Q8N3C5, Q8N9E9, Q8N9G2, Q96JL7 | Gene names | ABLIM2, KIAA1808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
ABLM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.132038 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BL65, Q6H8P8, Q80WK6, Q8BUT1, Q8BXI7 | Gene names | Ablim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
ABLM3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.130017 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
ABLM3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.130521 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q69ZX8, Q52KR1, Q6PAI7 | Gene names | Ablim3, Kiaa0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.064157 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CRIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.113477 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50238, Q13628, Q96J34 | Gene names | CRIP1, CRIP | |||
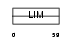
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich protein 1 (Cysteine-rich intestinal protein) (CRIP) (Cysteine-rich heart protein) (hCRHP). | |||||
|
CRIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.111741 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63254, P04006, Q3THF0 | Gene names | Crip1, Crip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich protein 1 (Cysteine-rich intestinal protein) (CRIP). | |||||
|
CRIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.110896 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P52943 | Gene names | CRIP2, CRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2 (CRP2) (Protein ESP1). | |||||
|
CRIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.112104 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9DCT8 | Gene names | Crip2, Crp2, Hlp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2 (CRP2) (Heart LIM protein). | |||||
|
CRIP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.111013 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6Q6R5, Q5T043, Q6Q6R4, Q6Q6R6, Q6Q6R7 | Gene names | CRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich protein 3 (CRP3) (Chromosome 6 LIM-only protein) (h6LIMo). | |||||
|
CRIP3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.086162 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6Q6R3, Q91V23, Q91V60 | Gene names | Crip3, Tlp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich protein 3 (CRP3) (Thymus LIM protein) (m17TLP). | |||||
|
FBLI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.159035 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
FHL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.173836 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13642, O95212, Q13230, Q13645, Q5M7Y6, Q9NZ40, Q9UKZ8, Q9Y630 | Gene names | FHL1, SLIM1 | |||
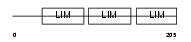
|
Domain Architecture |
|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM). | |||||
|
ISL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.095002 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61371, P20663, P47894 | Gene names | ISL1 | |||
|
Domain Architecture |
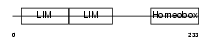
|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.095002 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61372, P20663, P47894, Q812D8 | Gene names | Isl1 | |||
|
Domain Architecture |
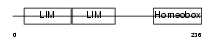
|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.096953 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |
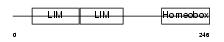
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
ISL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.096915 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CXV0 | Gene names | Isl2 | |||
|
Domain Architecture |
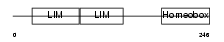
|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
LASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.069341 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14847, Q96ED2, Q96IG0 | Gene names | LASP1, MLN50 | |||
|
Domain Architecture |
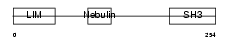
|
|||||
| Description | LIM and SH3 domain protein 1 (LASP-1) (MLN 50). | |||||
|
LASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.072205 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61792, Q62416 | Gene names | Lasp1, Mln50 | |||
|
Domain Architecture |
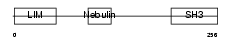
|
|||||
| Description | LIM and SH3 domain protein 1 (LASP-1) (MLN 50). | |||||
|
LHX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.088574 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48742, Q3MIW0 | Gene names | LHX1, LIM1 | |||
|
Domain Architecture |
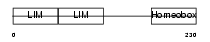
|
|||||
| Description | LIM/homeobox protein Lhx1 (LIM homeobox protein 1) (Homeobox protein Lim-1). | |||||
|
LHX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.088778 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P63006, P36199 | Gene names | Lhx1, Lim-1, Lim1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM/homeobox protein Lhx1 (LIM homeobox protein 1) (Homeobox protein Lim-1). | |||||
|
LHX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.091577 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |
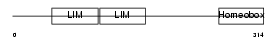
|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
LHX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.091091 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0S2 | Gene names | Lhx2 | |||
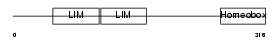
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx2. | |||||
|
LHX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.090727 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
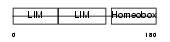
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
LHX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.090637 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
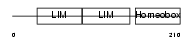
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
LHX4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.094115 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q969G2, Q8NHE0, Q8TCJ1, Q8WWX2, Q969W2 | Gene names | LHX4 | |||
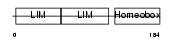
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
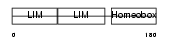
LHX4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.094098 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P53776, O08916, Q9R280 | Gene names | Lhx4, Gsh-4, Gsh4 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
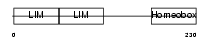
LHX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.087888 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2C1 | Gene names | LHX5 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
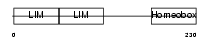
LHX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.088295 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
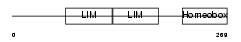
LHX61_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.093188 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UPM6, Q5T7S8, Q9NTK3, Q9UPM5 | Gene names | LHX6, LHX6.1 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx6.1 (Lhx6). | |||||
|
LHX61_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.096322 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R1R0, Q9R1R1 | Gene names | Lhx6.1 | |||
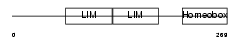
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx6.1. | |||||
|
LHX9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.092187 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQ69, Q5VUE3, Q9BYU6, Q9NQ70 | Gene names | LHX9 | |||
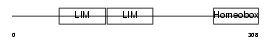
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
LHX9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.092230 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WUH2, Q9QYQ5, Q9QYQ6, Q9QZ00, Q9WU44 | Gene names | Lhx9 | |||
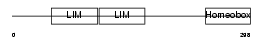
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
LIMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.073499 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
LIMA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.081172 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
LIMD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.134548 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.133531 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
LIMD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.084674 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BT23, Q96S91 | Gene names | LIMD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-containing protein 2. | |||||
|
LIMD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.084069 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGB5 | Gene names | Limd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-containing protein 2. | |||||
|
LMCD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.148601 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZU5 | Gene names | LMCD1 | |||
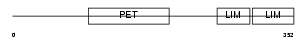
|
Domain Architecture |
|
|||||
| Description | LIM and cysteine-rich domains protein 1 (Dyxin). | |||||
|
LMCD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.143806 (rank : 49) | |||
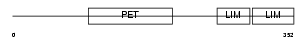
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VEE1 | Gene names | Lmcd1 | |||
|
Domain Architecture |
|
|||||
| Description | LIM and cysteine-rich domains protein 1. | |||||
|
LMX1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.102955 (rank : 75) | |||
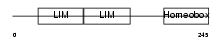
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TE12, Q8TE11 | Gene names | LMX1A | |||
|
Domain Architecture |
|
|||||
| Description | LIM homeobox transcription factor 1 alpha (LIM/homeobox protein LMX1A) (LIM-homeobox protein 1.1) (LMX-1.1). | |||||
|
LMX1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.105459 (rank : 70) | |||
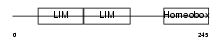
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JKU8 | Gene names | Lmx1a | |||
|
Domain Architecture |
|
|||||
| Description | LIM homeobox transcription factor 1 alpha (LIM/homeobox protein LMX1A) (LIM-homeobox protein 1.1) (LMX-1.1). | |||||
|
LMX1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.104437 (rank : 73) | |||
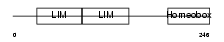
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60663, O75463 | Gene names | LMX1B | |||
|
Domain Architecture |
|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LMX1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.104606 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88609 | Gene names | Lmx1b | |||
|
Domain Architecture |
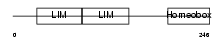
|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.116885 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
LPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.123168 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
LPXN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.157267 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99N69 | Gene names | Lpxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leupaxin. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.053370 (rank : 120) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
PAXI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.150571 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49023, O14970, O14971, O60360 | Gene names | PXN | |||
|
Domain Architecture |

|
|||||
| Description | Paxillin. | |||||
|
PAXI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.150585 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VI36, Q3TB62, Q3TZQ6, Q8VI37 | Gene names | Pxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paxillin. | |||||
|
PDLI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.069100 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |
|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
PDLI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.064905 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70400, Q99K93 | Gene names | Pdlim1, Clim1 | |||
|
Domain Architecture |
|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
PDLI3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.072538 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q53GG5, O43590, O60439, O60440, Q8N6Y6, Q9BVP4 | Gene names | PDLIM3, ALP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
PDLI3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.073469 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O70209 | Gene names | Pdlim3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
PDLI4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.075326 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50479, Q96AT8, Q9BTW8, Q9Y292 | Gene names | PDLIM4, RIL | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
PDLI4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.074995 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70271, Q8K0W4 | Gene names | Pdlim4, Ril | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
PINC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.156045 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48059, Q53TE0, Q9BS44 | Gene names | LIMS1 | |||
|
Domain Architecture |
|
|||||
| Description | Protein PINCH (Particularly interesting new Cys-His protein) (LIM and senescent cell antigen-like domains 1) (NY-REN-48 antigen). | |||||
|
RBTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.132334 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25791, Q9HD58 | Gene names | LMO2, RBTN2, RHOM2, TTG2 | |||
|
Domain Architecture |
|
|||||
| Description | Rhombotin-2 (Cysteine-rich protein TTG-2) (T-cell translocation protein 2) (LIM-only protein 2). | |||||
|
RBTN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.132338 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25801 | Gene names | Lmo2, Rbtn-2, Rbtn2, Rhom-2 | |||
|
Domain Architecture |
|
|||||
| Description | Rhombotin-2 (Cysteine-rich protein TTG-2) (T-cell translocation protein 2) (LIM-only protein 2). | |||||
|
TES_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.153241 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UGI8, Q9GZQ1, Q9HAJ9 | Gene names | TES | |||
|
Domain Architecture |
|
|||||
| Description | Testin (TESS). | |||||
|
TES_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.155838 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47226 | Gene names | Tes | |||
|
Domain Architecture |
|
|||||
| Description | Testin (TES1/TES2). | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.128086 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
ZYX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.143515 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
ZYX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.146094 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
ZN185_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | O15231, O00345, Q9NSD2 | Gene names | ZNF185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein ZNF185) (P1-A). | |||||
|
ZN185_MOUSE
|
||||||
| NC score | 0.894964 (rank : 2) | θ value | 1.07473e-77 (rank : 2) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62394 | Gene names | Znf185, Zfp185 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 185 (LIM-domain protein Zfp185) (P1-A). | |||||
|
SCEL_MOUSE
|
||||||
| NC score | 0.499291 (rank : 3) | θ value | 2.69671e-12 (rank : 3) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9EQG3, Q9CTT9 | Gene names | Scel | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
SCEL_HUMAN
|
||||||
| NC score | 0.492551 (rank : 4) | θ value | 4.59992e-12 (rank : 4) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O95171 | Gene names | SCEL | |||
|
Domain Architecture |

|
|||||
| Description | Sciellin. | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.261445 (rank : 5) | θ value | 3.77169e-06 (rank : 5) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
PRIC2_MOUSE
|
||||||
| NC score | 0.205991 (rank : 6) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q80Y24 | Gene names | Prickle2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
PRIC2_HUMAN
|
||||||
| NC score | 0.202573 (rank : 7) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z3G6 | Gene names | PRICKLE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 2. | |||||
|
CSRP1_MOUSE
|
||||||
| NC score | 0.200963 (rank : 8) | θ value | 0.21417 (rank : 27) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_HUMAN
|
||||||
| NC score | 0.200179 (rank : 9) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP2_MOUSE
|
||||||
| NC score | 0.200031 (rank : 10) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P97314, Q9CZG5 | Gene names | Csrp2, Dlp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1). | |||||
|
CSRP2_HUMAN
|
||||||
| NC score | 0.199934 (rank : 11) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q16527, Q93030 | Gene names | CSRP2, LMO5, SMLIM | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Smooth muscle cell LIM protein) (SmLIM) (LIM-only protein 5). | |||||
|
LMO6_MOUSE
|
||||||
| NC score | 0.199222 (rank : 12) | θ value | 0.0193708 (rank : 11) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q80VL3, Q8CAY9 | Gene names | Lmo6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
LMO6_HUMAN
|
||||||
| NC score | 0.197934 (rank : 13) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O43900, O76007 | Gene names | LMO6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 6 (Triple LIM domain protein 6). | |||||
|
FHL3_HUMAN
|
||||||
| NC score | 0.197139 (rank : 14) | θ value | 1.06291 (rank : 47) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q13643 | Gene names | FHL3, SLIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
PRIC1_HUMAN
|
||||||
| NC score | 0.193998 (rank : 15) | θ value | 0.365318 (rank : 37) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96MT3, Q71QF8, Q96N00 | Gene names | PRICKLE1, RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prickle-like protein 1 (REST/NRSF-interacting LIM domain protein 1). | |||||
|
FHL5_MOUSE
|
||||||
| NC score | 0.190258 (rank : 16) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9WTX7 | Gene names | Fhl5, Act | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
CSRP3_MOUSE
|
||||||
| NC score | 0.190214 (rank : 17) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50462 | Gene names | Csrp3, Clp, Mlp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 3 (Cysteine-rich protein 3) (CRP3) (LIM domain protein, cardiac) (Muscle LIM protein). | |||||
|
FHL3_MOUSE
|
||||||
| NC score | 0.189895 (rank : 18) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9R059, Q9JLP5, Q9WUH3 | Gene names | Fhl3 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
CSRP3_HUMAN
|
||||||
| NC score | 0.188382 (rank : 19) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P50461, Q9P131 | Gene names | CSRP3, CLP, MLP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 3 (Cysteine-rich protein 3) (CRP3) (LIM domain protein, cardiac) (Muscle LIM protein). | |||||
|
FHL2_HUMAN
|
||||||
| NC score | 0.179876 (rank : 20) | θ value | 0.0431538 (rank : 17) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 533 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q14192, Q13229, Q13644, Q9P294 | Gene names | FHL2, DRAL, SLIM3 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3) (LIM-domain protein DRAL). | |||||
|
FHL5_HUMAN
|
||||||
| NC score | 0.179055 (rank : 21) | θ value | 0.21417 (rank : 28) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q5TD97, Q5TD98, Q8WW21, Q9NQU2 | Gene names | FHL5, ACT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Four and a half LIM domains protein 5 (FHL-5) (Activator of cAMP- responsive element modulator in testis) (Activator of CREM in testis). | |||||
|
RBTN1_HUMAN
|
||||||
| NC score | 0.177587 (rank : 22) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P25800, Q4VBC5 | Gene names | LMO1, RBTN1, RHOM1, TTG1 | |||
|
Domain Architecture |

|
|||||
| Description | Rhombotin-1 (Cysteine-rich protein TTG-1) (T-cell translocation protein 1) (LIM-only protein 1). | |||||
|
RBTN1_MOUSE
|
||||||
| NC score | 0.177475 (rank : 23) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q924W9 | Gene names | Lmo1, Rbtn1, Rhom1, Ttg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rhombotin-1 (Cysteine-rich protein TTG-1) (LIM-only protein 1). | |||||
|
LMO3_HUMAN
|
||||||
| NC score | 0.176388 (rank : 24) | θ value | 0.00665767 (rank : 6) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8TAP4, Q8N974, Q9UDD5 | Gene names | LMO3, RBTN3, RHOM3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM-only protein 3 (Neuronal-specific transcription factor DAT1) (Rhombotin-3). | |||||
|
LMO3_MOUSE
|
||||||
| NC score | 0.176388 (rank : 25) | θ value | 0.00665767 (rank : 7) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BZL8 | Gene names | Lmo3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM-only protein 3 (Neuronal-specific transcription factor DAT1). | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.176279 (rank : 26) | θ value | 0.21417 (rank : 29) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.175159 (rank : 27) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
LDB3_MOUSE
|
||||||
| NC score | 0.174739 (rank : 28) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 510 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9JKS4, Q6A038, Q811P2, Q811P3, Q811P4, Q811P5, Q9D130, Q9JKS3, Q9R0Z1, Q9WVH1, Q9WVH2 | Gene names | Ldb3, Kiaa0613 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher) (Protein oracle). | |||||
|
FHL1_MOUSE
|
||||||
| NC score | 0.174540 (rank : 29) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 462 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97447, O55181, Q8K318 | Gene names | Fhl1 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM) (KyoT) (RBP-associated molecule 14-1) (RAM14-1). | |||||
|
FHL2_MOUSE
|
||||||
| NC score | 0.174423 (rank : 30) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O70433, P97448 | Gene names | Fhl2 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3). | |||||
|
PDLI5_MOUSE
|
||||||
| NC score | 0.174219 (rank : 31) | θ value | 0.163984 (rank : 24) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CI51, Q9QYN0, Q9QYN1, Q9QYN2 | Gene names | Pdlim5, Enh | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
FHL1_HUMAN
|
||||||
| NC score | 0.173836 (rank : 32) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13642, O95212, Q13230, Q13645, Q5M7Y6, Q9NZ40, Q9UKZ8, Q9Y630 | Gene names | FHL1, SLIM1 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 1 (FHL-1) (Skeletal muscle LIM- protein 1) (SLIM 1) (SLIM). | |||||
|
PDLI7_MOUSE
|
||||||
| NC score | 0.171176 (rank : 33) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3TJD7, Q80ZY6, Q810S3, Q8C1S4, Q9CRA1 | Gene names | Pdlim7, Enigma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
FBLI1_HUMAN
|
||||||
| NC score | 0.170889 (rank : 34) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8WUP2, Q5VVE0, Q5VVE1, Q8IX23, Q96T00, Q9UFD6 | Gene names | FBLIM1, FBLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-binding LIM protein 1 (Filamin-binding LIM protein 1) (FBLP-1) (Mitogen-inducible 2-interacting protein) (MIG2-interacting protein) (Migfilin). | |||||
|
CF049_HUMAN
|
||||||
| NC score | 0.169337 (rank : 35) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q2TBC4, Q5T3D4, Q9NSV1 | Gene names | C6orf49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf49. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.168184 (rank : 36) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
LPXN_HUMAN
|
||||||
| NC score | 0.161108 (rank : 37) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60711 | Gene names | LPXN, LDLP | |||
|
Domain Architecture |

|
|||||
| Description | Leupaxin. | |||||
|
FBLI1_MOUSE
|
||||||
| NC score | 0.159035 (rank : 38) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q71FD7, Q99J35 | Gene names | Fblim1, Cal | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-binding LIM protein 1 (CSX-associated LIM). | |||||
|
LPXN_MOUSE
|
||||||
| NC score | 0.157267 (rank : 39) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q99N69 | Gene names | Lpxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leupaxin. | |||||
|
PINC_HUMAN
|
||||||
| NC score | 0.156045 (rank : 40) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P48059, Q53TE0, Q9BS44 | Gene names | LIMS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein PINCH (Particularly interesting new Cys-His protein) (LIM and senescent cell antigen-like domains 1) (NY-REN-48 antigen). | |||||
|
TES_MOUSE
|
||||||
| NC score | 0.155838 (rank : 41) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P47226 | Gene names | Tes | |||
|
Domain Architecture |

|
|||||
| Description | Testin (TES1/TES2). | |||||
|
TES_HUMAN
|
||||||
| NC score | 0.153241 (rank : 42) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UGI8, Q9GZQ1, Q9HAJ9 | Gene names | TES | |||
|
Domain Architecture |

|
|||||
| Description | Testin (TESS). | |||||
|
PAXI_MOUSE
|
||||||
| NC score | 0.150585 (rank : 43) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VI36, Q3TB62, Q3TZQ6, Q8VI37 | Gene names | Pxn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paxillin. | |||||
|
PAXI_HUMAN
|
||||||
| NC score | 0.150571 (rank : 44) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P49023, O14970, O14971, O60360 | Gene names | PXN | |||
|
Domain Architecture |

|
|||||
| Description | Paxillin. | |||||
|
LMO4_HUMAN
|
||||||
| NC score | 0.149226 (rank : 45) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61968, O00158, O88894 | Gene names | LMO4 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain transcription factor LMO4 (LIM domain only protein 4) (LMO- 4) (Breast tumor autoantigen). | |||||
|
LMO4_MOUSE
|
||||||
| NC score | 0.149226 (rank : 46) | θ value | 2.36792 (rank : 65) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61969, O00158, O88894 | Gene names | Lmo4 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain transcription factor LMO4 (LIM domain only protein 4) (LMO- 4) (Breast tumor autoantigen). | |||||
|
LMCD1_HUMAN
|
||||||
| NC score | 0.148601 (rank : 47) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZU5 | Gene names | LMCD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM and cysteine-rich domains protein 1 (Dyxin). | |||||
|
ZYX_MOUSE
|
||||||
| NC score | 0.146094 (rank : 48) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
LMCD1_MOUSE
|
||||||
| NC score | 0.143806 (rank : 49) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8VEE1 | Gene names | Lmcd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM and cysteine-rich domains protein 1. | |||||
|
ZYX_HUMAN
|
||||||
| NC score | 0.143515 (rank : 50) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q15942 | Gene names | ZYX | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin (Zyxin-2). | |||||
|
LIMD1_HUMAN
|
||||||
| NC score | 0.134548 (rank : 51) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UGP4, Q9BQQ9, Q9NQ47 | Gene names | LIMD1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
ABLM1_MOUSE
|
||||||
| NC score | 0.134003 (rank : 52) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8K4G5, Q80U86, Q8BIR9, Q8K4G3, Q8K4G4 | Gene names | Ablim1, Ablim, Kiaa0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1). | |||||
|
ABLM1_HUMAN
|
||||||
| NC score | 0.133582 (rank : 53) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O14639, Q15039, Q68CQ9, Q9BUP1 | Gene names | ABLIM1, ABLIM, KIAA0059 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 1 (Actin-binding LIM protein family member 1) (abLIM-1) (Actin-binding double-zinc-finger protein) (LIMAB1) (Limatin). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.133531 (rank : 54) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
RBTN2_MOUSE
|
||||||
| NC score | 0.132338 (rank : 55) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25801 | Gene names | Lmo2, Rbtn-2, Rbtn2, Rhom-2 | |||
|
Domain Architecture |

|
|||||
| Description | Rhombotin-2 (Cysteine-rich protein TTG-2) (T-cell translocation protein 2) (LIM-only protein 2). | |||||
|
RBTN2_HUMAN
|
||||||
| NC score | 0.132334 (rank : 56) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25791, Q9HD58 | Gene names | LMO2, RBTN2, RHOM2, TTG2 | |||
|
Domain Architecture |

|
|||||
| Description | Rhombotin-2 (Cysteine-rich protein TTG-2) (T-cell translocation protein 2) (LIM-only protein 2). | |||||
|
ABLM2_MOUSE
|
||||||
| NC score | 0.132038 (rank : 57) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BL65, Q6H8P8, Q80WK6, Q8BUT1, Q8BXI7 | Gene names | Ablim2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
ABLM2_HUMAN
|
||||||
| NC score | 0.130875 (rank : 58) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6H8Q1, Q6H8Q0, Q6NX73, Q8N3C5, Q8N9E9, Q8N9G2, Q96JL7 | Gene names | ABLIM2, KIAA1808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 2 (Actin-binding LIM protein family member 2) (abLIM-2). | |||||
|
ABLM3_MOUSE
|
||||||
| NC score | 0.130521 (rank : 59) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q69ZX8, Q52KR1, Q6PAI7 | Gene names | Ablim3, Kiaa0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
ABLM3_HUMAN
|
||||||
| NC score | 0.130017 (rank : 60) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O94929, Q658S1, Q68CI5, Q9BV32 | Gene names | ABLIM3, KIAA0843 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding LIM protein 3 (Actin-binding LIM protein family member 3) (abLIM-3). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.128086 (rank : 61) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
LPP_MOUSE
|
||||||
| NC score | 0.123168 (rank : 62) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BFW7, Q5U407, Q8BHI1, Q8BKI0, Q8BKN2, Q8BLF4, Q8BLG3, Q8C101 | Gene names | Lpp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner homolog. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.116885 (rank : 63) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CRIP1_HUMAN
|
||||||
| NC score | 0.113477 (rank : 64) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P50238, Q13628, Q96J34 | Gene names | CRIP1, CRIP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 1 (Cysteine-rich intestinal protein) (CRIP) (Cysteine-rich heart protein) (hCRHP). | |||||
|
CRIP2_MOUSE
|
||||||
| NC score | 0.112104 (rank : 65) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9DCT8 | Gene names | Crip2, Crp2, Hlp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2 (CRP2) (Heart LIM protein). | |||||
|
CRIP1_MOUSE
|
||||||
| NC score | 0.111741 (rank : 66) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P63254, P04006, Q3THF0 | Gene names | Crip1, Crip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich protein 1 (Cysteine-rich intestinal protein) (CRIP). | |||||
|
CRIP3_HUMAN
|
||||||
| NC score | 0.111013 (rank : 67) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q6Q6R5, Q5T043, Q6Q6R4, Q6Q6R6, Q6Q6R7 | Gene names | CRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich protein 3 (CRP3) (Chromosome 6 LIM-only protein) (h6LIMo). | |||||
|
CRIP2_HUMAN
|
||||||
| NC score | 0.110896 (rank : 68) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P52943 | Gene names | CRIP2, CRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich protein 2 (CRP2) (Protein ESP1). | |||||
|
LHX8_MOUSE
|
||||||
| NC score | 0.107047 (rank : 69) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O35652, O70163, O88707 | Gene names | Lhx8, Lhx7 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx8 (LIM homeodomain Lhx7) (L3). | |||||
|
LMX1A_MOUSE
|
||||||
| NC score | 0.105459 (rank : 70) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9JKU8 | Gene names | Lmx1a | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 alpha (LIM/homeobox protein LMX1A) (LIM-homeobox protein 1.1) (LMX-1.1). | |||||
|
LHX6_MOUSE
|
||||||
| NC score | 0.104618 (rank : 71) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88706 | Gene names | Lhx6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6. | |||||
|
LMX1B_MOUSE
|
||||||
| NC score | 0.104606 (rank : 72) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88609 | Gene names | Lmx1b | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
LMX1B_HUMAN
|
||||||
| NC score | 0.104437 (rank : 73) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O60663, O75463 | Gene names | LMX1B | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 beta (LIM/homeobox protein LMX1B) (LIM-homeobox protein 1.2) (LMX-1.2). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.103048 (rank : 74) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
LMX1A_HUMAN
|
||||||
| NC score | 0.102955 (rank : 75) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8TE12, Q8TE11 | Gene names | LMX1A | |||
|
Domain Architecture |

|
|||||
| Description | LIM homeobox transcription factor 1 alpha (LIM/homeobox protein LMX1A) (LIM-homeobox protein 1.1) (LMX-1.1). | |||||
|
ISL2_HUMAN
|
||||||
| NC score | 0.096953 (rank : 76) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q96A47 | Gene names | ISL2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
ISL2_MOUSE
|
||||||
| NC score | 0.096915 (rank : 77) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9CXV0 | Gene names | Isl2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-2 (Islet-2). | |||||
|
LHX61_MOUSE
|
||||||
| NC score | 0.096322 (rank : 78) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9R1R0, Q9R1R1 | Gene names | Lhx6.1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6.1. | |||||
|
ISL1_HUMAN
|
||||||
| NC score | 0.095002 (rank : 79) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61371, P20663, P47894 | Gene names | ISL1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
ISL1_MOUSE
|
||||||
| NC score | 0.095002 (rank : 80) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61372, P20663, P47894, Q812D8 | Gene names | Isl1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin gene enhancer protein ISL-1 (Islet-1). | |||||
|
LHX4_HUMAN
|
||||||
| NC score | 0.094115 (rank : 81) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q969G2, Q8NHE0, Q8TCJ1, Q8WWX2, Q969W2 | Gene names | LHX4 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
LHX4_MOUSE
|
||||||
| NC score | 0.094098 (rank : 82) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P53776, O08916, Q9R280 | Gene names | Lhx4, Gsh-4, Gsh4 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx4. | |||||
|
LHX61_HUMAN
|
||||||
| NC score | 0.093188 (rank : 83) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UPM6, Q5T7S8, Q9NTK3, Q9UPM5 | Gene names | LHX6, LHX6.1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6.1 (Lhx6). | |||||
|
LHX9_MOUSE
|
||||||
| NC score | 0.092230 (rank : 84) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9WUH2, Q9QYQ5, Q9QYQ6, Q9QZ00, Q9WU44 | Gene names | Lhx9 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
LHX9_HUMAN
|
||||||
| NC score | 0.092187 (rank : 85) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9NQ69, Q5VUE3, Q9BYU6, Q9NQ70 | Gene names | LHX9 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx9. | |||||
|
LHX2_HUMAN
|
||||||
| NC score | 0.091577 (rank : 86) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
LHX2_MOUSE
|
||||||
| NC score | 0.091091 (rank : 87) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z0S2 | Gene names | Lhx2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2. | |||||
|
LHX3_HUMAN
|
||||||
| NC score | 0.090727 (rank : 88) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9UBR4, Q9NZB5, Q9P0I8, Q9P0I9 | Gene names | LHX3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3. | |||||
|
LHX3_MOUSE
|
||||||
| NC score | 0.090637 (rank : 89) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50481, Q61800, Q61801 | Gene names | Lhx3, Lim-3, Lim3, Plim | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx3 (Homeobox protein LIM-3) (Homeobox protein P-LIM) (LIM homeobox protein 3). | |||||
|
LHX1_MOUSE
|
||||||
| NC score | 0.088778 (rank : 90) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P63006, P36199 | Gene names | Lhx1, Lim-1, Lim1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM/homeobox protein Lhx1 (LIM homeobox protein 1) (Homeobox protein Lim-1). | |||||
|
LHX1_HUMAN
|
||||||
| NC score | 0.088574 (rank : 91) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48742, Q3MIW0 | Gene names | LHX1, LIM1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx1 (LIM homeobox protein 1) (Homeobox protein Lim-1). | |||||
|
LHX5_MOUSE
|
||||||
| NC score | 0.088295 (rank : 92) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
LHX5_HUMAN
|
||||||
| NC score | 0.087888 (rank : 93) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H2C1 | Gene names | LHX5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
CJ092_HUMAN
|
||||||
| NC score | 0.086936 (rank : 94) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IYW2, Q5JSF7, Q9NTQ5 | Gene names | C10orf92 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf92. | |||||
|
CRIP3_MOUSE
|
||||||
| NC score | 0.086162 (rank : 95) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6Q6R3, Q91V23, Q91V60 | Gene names | Crip3, Tlp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich protein 3 (CRP3) (Thymus LIM protein) (m17TLP). | |||||
|
LIMD2_HUMAN
|
||||||
| NC score | 0.084674 (rank : 96) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9BT23, Q96S91 | Gene names | LIMD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-containing protein 2. | |||||
|
SF3B1_MOUSE
|
||||||
| NC score | 0.084282 (rank : 97) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99NB9, Q9CSK5 | Gene names | Sf3b1, Sap155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
LIMD2_MOUSE
|
||||||
| NC score | 0.084069 (rank : 98) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGB5 | Gene names | Limd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-containing protein 2. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.082568 (rank : 99) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
LIMA1_MOUSE
|
||||||
| NC score | 0.081172 (rank : 100) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9ERG0, Q9ERG1 | Gene names | Lima1, D15Ertd366e, Eplin | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm) (mEPLIN). | |||||
|
SF3B1_HUMAN
|
||||||
| NC score | 0.080640 (rank : 101) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75533 | Gene names | SF3B1, SAP155 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 1 (Spliceosome-associated protein 155) (SAP 155) (SF3b155) (Pre-mRNA-splicing factor SF3b 155 kDa subunit). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.078797 (rank : 102) | θ value | 0.0330416 (rank : 16) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
PDLI4_HUMAN
|
||||||
| NC score | 0.075326 (rank : 103) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P50479, Q96AT8, Q9BTW8, Q9Y292 | Gene names | PDLIM4, RIL | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
PDLI4_MOUSE
|
||||||
| NC score | 0.074995 (rank : 104) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70271, Q8K0W4 | Gene names | Pdlim4, Ril | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 4 (LIM protein RIL) (Reversion-induced LIM protein). | |||||
|
LIMA1_HUMAN
|
||||||
| NC score | 0.073499 (rank : 105) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9UHB6, Q2TAN7, Q9BVF2, Q9H8J1, Q9HBN5, Q9NX96, Q9NXC3, Q9NXU6, Q9P0H8, Q9UHB5 | Gene names | LIMA1, EPLIN, SREBP3 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain and actin-binding protein 1 (Epithelial protein lost in neoplasm). | |||||
|
PDLI3_MOUSE
|
||||||
| NC score | 0.073469 (rank : 106) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O70209 | Gene names | Pdlim3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
PDLI3_HUMAN
|
||||||
| NC score | 0.072538 (rank : 107) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q53GG5, O43590, O60439, O60440, Q8N6Y6, Q9BVP4 | Gene names | PDLIM3, ALP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 3 (Actinin-associated LIM protein) (Alpha- actinin-2-associated LIM protein). | |||||
|
LASP1_MOUSE
|
||||||
| NC score | 0.072205 (rank : 108) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61792, Q62416 | Gene names | Lasp1, Mln50 | |||
|
Domain Architecture |

|
|||||
| Description | LIM and SH3 domain protein 1 (LASP-1) (MLN 50). | |||||
|
LASP1_HUMAN
|
||||||
| NC score | 0.069341 (rank : 109) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14847, Q96ED2, Q96IG0 | Gene names | LASP1, MLN50 | |||
|
Domain Architecture |

|
|||||
| Description | LIM and SH3 domain protein 1 (LASP-1) (MLN 50). | |||||
|
PDLI1_HUMAN
|
||||||
| NC score | 0.069100 (rank : 110) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O00151, Q5VZH5, Q9BPZ9 | Gene names | PDLIM1, CLIM1, CLP36 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.067458 (rank : 111) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
CLOCK_HUMAN
|
||||||
| NC score | 0.065451 (rank : 112) | θ value | 0.0563607 (rank : 19) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15516, O14516, Q9UIT8 | Gene names | CLOCK, KIAA0334 | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (hCLOCK). | |||||
|
PDLI1_MOUSE
|
||||||
| NC score | 0.064905 (rank : 113) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O70400, Q99K93 | Gene names | Pdlim1, Clim1 | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 1 (Elfin) (LIM domain protein CLP-36) (C- terminal LIM domain protein 1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.064157 (rank : 114) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.061538 (rank : 115) | θ value | 0.21417 (rank : 30) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.060854 (rank : 116) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.056821 (rank : 117) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.055742 (rank : 118) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.053789 (rank : 119) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.053370 (rank : 120) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.051578 (rank : 121) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.050608 (rank : 122) | θ value | 4.03905 (rank : 81) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.050118 (rank : 123) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MAP4_MOUSE
|
||||||
| NC score | 0.048576 (rank : 124) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P27546 | Gene names | Map4, Mtap4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.048116 (rank : 125) | θ value | 6.88961 (rank : 102) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.047318 (rank : 126) | θ value | 1.06291 (rank : 48) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.045293 (rank : 127) | θ value | 0.0148317 (rank : 10) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BBX_MOUSE
|
||||||
| NC score | 0.044444 (rank : 128) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VBW5, Q3TZK1, Q6NXY8, Q6PEU3, Q8BQJ7, Q8C7E0, Q8CDQ0, Q8CDV1, Q8VI48, Q8VI49, Q8VI50, Q9CS94 | Gene names | Bbx, Hbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HMG box transcription factor BBX (Bobby sox homolog) (HMG box- containing protein 2). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.044215 (rank : 129) | θ value | 1.06291 (rank : 46) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.043749 (rank : 130) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.043449 (rank : 131) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.043428 (rank : 132) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.043100 (rank : 133) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CG1_HUMAN
|
||||||
| NC score | 0.041361 (rank : 134) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13495 | Gene names | CXorf6, CG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CG1 protein (F18). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.041031 (rank : 135) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
ISK6_HUMAN
|
||||||
| NC score | 0.040545 (rank : 136) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UWN8, Q8N5P0 | Gene names | SPINK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease inhibitor Kazal-type 6 precursor. | |||||
|
LIMK1_MOUSE
|
||||||
| NC score | 0.040291 (rank : 137) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53668 | Gene names | Limk1, Limk | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain kinase 1 (EC 2.7.11.1) (LIMK-1) (KIZ-1). | |||||
|
ZN488_HUMAN
|
||||||
| NC score | 0.039515 (rank : 138) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96MN9 | Gene names | ZNF488 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 488. | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.035702 (rank : 139) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.034712 (rank : 140) | θ value | 2.36792 (rank : 67) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.033187 (rank : 141) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.032483 (rank : 142) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.032245 (rank : 143) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.031878 (rank : 144) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.029578 (rank : 145) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.027128 (rank : 146) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
TBX15_HUMAN
|
||||||
| NC score | 0.026484 (rank : 147) | θ value | 2.36792 (rank : 66) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.026340 (rank : 148) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.026181 (rank : 149) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.025671 (rank : 150) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CDT1_HUMAN
|
||||||
| NC score | 0.025654 (rank : 151) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H211, Q86XX9, Q96CJ5, Q96GK5, Q96H67, Q96HE6, Q9BWM0 | Gene names | CDT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA replication factor Cdt1 (Double parked homolog) (DUP). | |||||
|
LAX1_MOUSE
|
||||||
| NC score | 0.024584 (rank : 152) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BHB3, Q8BS18, Q8CGV1 | Gene names | Lax1, Lax | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lymphocyte transmembrane adapter 1 (Membrane-associated adapter protein LAX) (Linker for activation of X cells). | |||||
|
LIPB2_MOUSE
|
||||||
| NC score | 0.024056 (rank : 153) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35711, Q7TMG4, Q8CBS6, Q99KX6 | Gene names | Ppfibp2, Cclp1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 2) (PTPRF-interacting protein-binding protein 2) (Coiled-coil-like protein 1). | |||||
|
SOX30_MOUSE
|
||||||
| NC score | 0.022291 (rank : 154) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CGW4 | Gene names | Sox30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
CO039_MOUSE
|
||||||
| NC score | 0.021638 (rank : 155) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3TEI4, Q3TMF2, Q3U253, Q3UM75, Q9DA93 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C15orf39 homolog. | |||||
|
ZN447_HUMAN
|
||||||
| NC score | 0.020306 (rank : 156) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TBC5, Q9BRK7, Q9H9A0 | Gene names | ZNF447 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 447. | |||||
|
TSSC4_HUMAN
|
||||||
| NC score | 0.019024 (rank : 157) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5U2, Q86VL2, Q9BRS6 | Gene names | TSSC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4 (Tumor-suppressing subchromosomal transferable fragment candidate gene 4 protein) (Tumor-suppressing STF cDNA 4 protein). | |||||
|
SOX13_MOUSE
|
||||||
| NC score | 0.016211 (rank : 158) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 495 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q04891, Q922L3 | Gene names | Sox13, Sox-13 | |||
|
Domain Architecture |

|
|||||
| Description | SOX-13 protein. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.016190 (rank : 159) | θ value | 6.88961 (rank : 103) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CBX2_MOUSE
|
||||||
| NC score | 0.016038 (rank : 160) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30658, O35731, Q8CIA0 | Gene names | Cbx2, M33 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 2 (Modifier 3 protein) (M33). | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.015978 (rank : 161) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
NGN2_MOUSE
|
||||||
| NC score | 0.015126 (rank : 162) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70447, P70237 | Gene names | Neurog2, Ath4a, Atoh4, Ngn2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenin 2 (Atonal protein homolog 4) (Helix-loop-helix protein mATH-4A) (MATH4A). | |||||
|
S4A7_HUMAN
|
||||||
| NC score | 0.014919 (rank : 163) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6M7, O60350, Q6AHZ9, Q9HC88, Q9UIB9 | Gene names | SLC4A7, BT, NBC2, NBC2B, NBC3, SBC2, SLC4A6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium bicarbonate cotransporter 3 (Sodium bicarbonate cotransporter 2) (Sodium bicarbonate cotransporter 2b) (Bicarbonate transporter) (Solute carrier family 4 member 7). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.014789 (rank : 164) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SC10A_HUMAN
|
||||||
| NC score | 0.014716 (rank : 165) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5Y9 | Gene names | SCN10A, PN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 10 subunit alpha (Sodium channel protein type X subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.8) (Peripheral nerve sodium channel 3) (hPN3). | |||||
|
CHSS3_HUMAN
|
||||||
| NC score | 0.014578 (rank : 166) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q70JA7, Q76L22, Q86Y52 | Gene names | CSS3, CHSY2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate synthase 3 (EC 2.4.1.175) (Glucuronosyl-N- acetylgalactosaminyl-proteoglycan 4-beta-N- acetylgalactosaminyltransferase II) (Chondroitin synthase 2) (N- acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase II) (EC 2.4.1.226) (Chondroitin glucuronyltransferase II) (N- acetylgalactosaminyltransferase II) (Carbohydrate synthase 2). | |||||
|
UBP34_HUMAN
|
||||||
| NC score | 0.014403 (rank : 167) | θ value | 4.03905 (rank : 82) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q70CQ2, O60316, O94834, Q3B777, Q6P6C9, Q7L8P6, Q8N3T9, Q8TBW2, Q9UGA1 | Gene names | USP34, KIAA0570, KIAA0729 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 34 (EC 3.1.2.15) (Ubiquitin thioesterase 34) (Ubiquitin-specific-processing protease 34) (Deubiquitinating enzyme 34). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.012383 (rank : 168) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.012100 (rank : 169) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
SALL2_MOUSE
|
||||||
| NC score | 0.011001 (rank : 170) | θ value | 6.88961 (rank : 104) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QX96 | Gene names | Sall2, Sal2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Spalt-like protein 2) (MSal-2). | |||||
|
ZN553_MOUSE
|
||||||
| NC score | 0.009303 (rank : 171) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3US17, Q3UFP3, Q8R0V0, Q921H7 | Gene names | Znf553, Zfp553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 553. | |||||
|
ZN553_HUMAN
|
||||||
| NC score | 0.009200 (rank : 172) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96MX3, Q4G0R3, Q69YP3, Q96IL9 | Gene names | ZNF553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 553. | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.007929 (rank : 173) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
ZN271_HUMAN
|
||||||
| NC score | 0.007831 (rank : 174) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14591, Q96T29, Q9BSX2, Q9UN33, Q9Y5B7 | Gene names | ZNF271, ZNFEB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 271 (Zinc finger protein HZF7) (Zinc finger protein ZNFphex133) (Epstein-Barr virus-induced zinc finger protein) (ZNF-EB) (CT-ZFP48) (Zinc finger protein dp) (ZNF-dp). | |||||
|
ZN181_HUMAN
|
||||||
| NC score | 0.005767 (rank : 175) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q2M3W8, Q49A75 | Gene names | ZNF181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 181 (HHZ181). | |||||
|
BCL6B_HUMAN
|
||||||
| NC score | 0.005580 (rank : 176) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 855 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N143, Q6PCB4 | Gene names | BCL6B, BAZF, ZNF62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell CLL/lymphoma 6 member B protein (Bcl6-associated zinc finger protein) (Zinc finger protein 62). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.000434 (rank : 177) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 112 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||