Please be patient as the page loads
|
SETB1_HUMAN
|
||||||
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SETB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.977838 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB2_HUMAN
|
||||||
| θ value | 1.96772e-63 (rank : 3) | NC score | 0.855961 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 4) | NC score | 0.356773 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_MOUSE
|
||||||
| θ value | 4.01107e-24 (rank : 5) | NC score | 0.353924 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 1.16704e-23 (rank : 6) | NC score | 0.348462 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
SUV91_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 7) | NC score | 0.652258 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 1.33526e-19 (rank : 8) | NC score | 0.643508 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_MOUSE
|
||||||
| θ value | 1.80466e-16 (rank : 9) | NC score | 0.625564 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 10) | NC score | 0.625135 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 3.89403e-11 (rank : 11) | NC score | 0.428437 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
SETD2_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 12) | NC score | 0.518046 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 13) | NC score | 0.423731 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 14) | NC score | 0.333410 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 15) | NC score | 0.311801 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 3.08544e-08 (rank : 16) | NC score | 0.426938 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 17) | NC score | 0.424029 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 18) | NC score | 0.323580 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 19) | NC score | 0.458273 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | 2.21117e-06 (rank : 20) | NC score | 0.469558 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 21) | NC score | 0.469129 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 22) | NC score | 0.464149 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
EZH2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 23) | NC score | 0.456659 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 24) | NC score | 0.074397 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 25) | NC score | 0.065120 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
HN1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.071729 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97825 | Gene names | Hn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1 protein. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 27) | NC score | 0.077205 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
WDR7_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 28) | NC score | 0.034579 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E6, Q86UX5, Q86VP2, Q96PS7 | Gene names | WDR7, KIAA0541, TRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 7 (TGF-beta resistance-associated protein TRAG) (Rabconnectin-3 beta). | |||||
|
FIBA_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 29) | NC score | 0.025367 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
F126A_MOUSE
|
||||||
| θ value | 0.163984 (rank : 30) | NC score | 0.057787 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P9N1, Q99NC4 | Gene names | Fam126a, Drctnnb1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM126A (Down-regulated by CTNNB1 protein A). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 31) | NC score | 0.041685 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.21417 (rank : 32) | NC score | 0.077614 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CD2L7_HUMAN
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.005506 (rank : 160) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | 0.279714 (rank : 34) | NC score | 0.022926 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
CHD2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.027651 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
EXTL1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 36) | NC score | 0.025411 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKV7 | Gene names | Extl1 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 1 (EC 2.4.1.224) (Glucuronosyl-N-acetylglucosaminyl- proteoglycan 4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L) (Multiple exostosis-like protein). | |||||
|
AKAP2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.054119 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.046410 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
FOXD1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.011670 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.024088 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
OR2J1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 41) | NC score | 0.002424 (rank : 166) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZK6, Q9GZK1 | Gene names | OR2J1 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 2J1 (Olfactory receptor 6-5) (OR6-5) (Hs6M1-4). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.020631 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
RN165_HUMAN
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.029248 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.033216 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.026271 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.028023 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.021516 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.043659 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 49) | NC score | 0.057768 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.046073 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
EP400_MOUSE
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.033259 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
MYCD_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.041987 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
ZF106_HUMAN
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.029362 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.030156 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
HS74L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.015684 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
RASF8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.038936 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CJ96, Q8CCW4, Q9CU91 | Gene names | Rassf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1 homolog). | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.024646 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CNGB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.016958 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CT055_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.041202 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R184, Q8BQE5, Q8K024, Q8VEM7, Q9CU80 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55 homolog. | |||||
|
CT2NL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 60) | NC score | 0.036909 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
ETV4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 61) | NC score | 0.011065 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.028679 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.015576 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.050637 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.032518 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.031281 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.007669 (rank : 156) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
EP15R_MOUSE
|
||||||
| θ value | 2.36792 (rank : 68) | NC score | 0.028432 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
F126A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 69) | NC score | 0.044593 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYI3, Q6N010, Q75MR4, Q7LDZ4, Q96MX1, Q96NQ6 | Gene names | FAM126A, DRCTNNB1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM126A (Down-regulated by CTNNB1 protein A). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 70) | NC score | 0.037095 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.014487 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.044956 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FLNB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.023677 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.042521 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LRCH3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 75) | NC score | 0.006578 (rank : 157) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 76) | NC score | 0.065561 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 77) | NC score | 0.048276 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
NTNG1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 78) | NC score | 0.015379 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
OR2J3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 79) | NC score | 0.000259 (rank : 167) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76001, Q96R15, Q9GZK5, Q9GZL4, Q9GZL5 | Gene names | OR2J3 | |||
|
Domain Architecture |
|
|||||
| Description | Olfactory receptor 2J3 (Olfactory receptor 6-6) (OR6-6) (Hs6M1-3). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 3.0926 (rank : 80) | NC score | 0.035290 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 81) | NC score | 0.012821 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TACC2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 82) | NC score | 0.029667 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
BLNK_MOUSE
|
||||||
| θ value | 4.03905 (rank : 83) | NC score | 0.029350 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 84) | NC score | 0.027269 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
DDX58_MOUSE
|
||||||
| θ value | 4.03905 (rank : 85) | NC score | 0.011772 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 86) | NC score | 0.047541 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
K1543_MOUSE
|
||||||
| θ value | 4.03905 (rank : 87) | NC score | 0.041683 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
NCOA7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 88) | NC score | 0.014936 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
PITM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 89) | NC score | 0.011094 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 90) | NC score | 0.030530 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SELS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 91) | NC score | 0.025346 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQE4, Q3B771, Q9P0I6 | Gene names | SELS, VIMP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein). | |||||
|
SEM6B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 92) | NC score | 0.006075 (rank : 158) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 93) | NC score | 0.038329 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
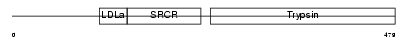
TMPS2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 94) | NC score | 0.004050 (rank : 164) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TSC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.026774 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
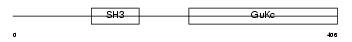
CACB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 96) | NC score | 0.012399 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02641, O15331, Q02639, Q02640, Q9C085 | Gene names | CACNB1, CACNLB1 | |||
|
Domain Architecture |
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 97) | NC score | 0.012349 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3Z5, O88517, Q7TPF2, Q8R3Z6, Q9EPT9 | Gene names | Cacnb1, Cacnlb1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
FOXO1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 98) | NC score | 0.008580 (rank : 155) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12778, O43523, Q5VYC7, Q6NSK6 | Gene names | FOXO1A, FKHR | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 99) | NC score | 0.024345 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 100) | NC score | 0.010368 (rank : 151) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 101) | NC score | 0.009678 (rank : 152) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 102) | NC score | 0.029266 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 103) | NC score | 0.021097 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SSH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 104) | NC score | 0.011887 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.027849 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.005102 (rank : 161) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
WTAP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.028265 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15007, Q96T28, Q9BYJ7, Q9H4E2 | Gene names | WTAP, KIAA0105 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.006058 (rank : 159) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
ZIC3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | -0.000886 (rank : 169) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60481 | Gene names | ZIC3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein ZIC 3 (Zinc finger protein of the cerebellum 3). | |||||
|
ZIC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | -0.000880 (rank : 168) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62521 | Gene names | Zic3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein ZIC 3 (Zinc finger protein of the cerebellum 3). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | -0.001216 (rank : 170) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ATRX_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.026477 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.016009 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.026197 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
KRA11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.010615 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
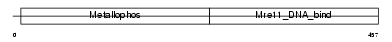
MRE11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 116) | NC score | 0.013451 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49959, O43475 | Gene names | MRE11A, HNGS1, MRE11 | |||
|
Domain Architecture |
|
|||||
| Description | Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A). | |||||
|
PALB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 117) | NC score | 0.014749 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
POK3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 118) | NC score | 0.004817 (rank : 162) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WJR5, O15312 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pol protein (HERV-K(C19) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 119) | NC score | 0.018767 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 120) | NC score | 0.018794 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SON_HUMAN
|
||||||
| θ value | 6.88961 (rank : 121) | NC score | 0.044999 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
UT14A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.019985 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
VMDL3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.009082 (rank : 153) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
VPS72_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.060362 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
CENPJ_HUMAN
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.018998 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.022308 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.037485 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 128) | NC score | 0.022771 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
JAG1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 129) | NC score | 0.008836 (rank : 154) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 130) | NC score | 0.017859 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
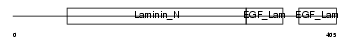
NTNG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 131) | NC score | 0.010918 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
PDIP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 132) | NC score | 0.011016 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG81, Q6PE83, Q80X99, Q8CI28 | Gene names | Poldip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase delta-interacting protein 3. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 133) | NC score | 0.027648 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PZRN4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 134) | NC score | 0.004150 (rank : 163) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
RNF17_HUMAN
|
||||||
| θ value | 8.99809 (rank : 135) | NC score | 0.012245 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXT8, Q9BXT7 | Gene names | RNF17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 136) | NC score | 0.027178 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 137) | NC score | 0.026780 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TRI62_MOUSE
|
||||||
| θ value | 8.99809 (rank : 138) | NC score | 0.003985 (rank : 165) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80V85 | Gene names | Trim62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 62. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.022894 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.012674 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
ZN445_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | -0.002917 (rank : 171) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59923 | Gene names | ZNF445, ZNF168 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 445. | |||||
|
ANR22_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.051178 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.063541 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.063541 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.064540 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.065174 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.061462 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.060845 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.050501 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.054793 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.061777 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.063004 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050644 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.050838 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.062863 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.061377 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.084712 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.064470 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.062772 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PHF7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.050819 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.062551 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.066741 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.051985 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SETD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.057832 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
SETD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.058870 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.308985 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
SETD8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.310906 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.050582 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
TARSH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.053273 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.062082 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.056976 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
SETB1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q15047, Q96GM9 | Gene names | SETDB1, KIAA0067 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB1_MOUSE
|
||||||
| NC score | 0.977838 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O88974, Q922K1 | Gene names | Setdb1, Eset | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 4 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 4) (H3-K9-HMTase 4) (SET domain bifurcated 1) (ERG-associated protein with SET domain) (ESET). | |||||
|
SETB2_HUMAN
|
||||||
| NC score | 0.855961 (rank : 3) | θ value | 1.96772e-63 (rank : 3) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96T68, Q86UD6, Q96AI6 | Gene names | SETDB2, CLLD8 | |||
|
Domain Architecture |

|
|||||
| Description | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific (EC 2.1.1.43) (Histone H3-K9 methyltransferase) (H3-K9-HMTase) (SET domain bifurcated 2) (Chronic lymphocytic leukemia deletion region gene 8 protein). | |||||
|
SUV91_HUMAN
|
||||||
| NC score | 0.652258 (rank : 4) | θ value | 1.5773e-20 (rank : 7) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O43463, Q53G60, Q6FHK6 | Gene names | SUV39H1, SUV39H | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.643508 (rank : 5) | θ value | 1.33526e-19 (rank : 8) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
SUV92_MOUSE
|
||||||
| NC score | 0.625564 (rank : 6) | θ value | 1.80466e-16 (rank : 9) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9EQQ0, Q9CUK3, Q9JLP7 | Gene names | Suv39h2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SUV92_HUMAN
|
||||||
| NC score | 0.625135 (rank : 7) | θ value | 3.40345e-15 (rank : 10) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9H5I1 | Gene names | SUV39H2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 2 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 2) (H3-K9-HMTase 2) (Suppressor of variegation 3-9 homolog 2) (Su(var)3-9 homolog 2). | |||||
|
SETD2_HUMAN
|
||||||
| NC score | 0.518046 (rank : 8) | θ value | 1.9326e-10 (rank : 12) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9BYW2, O75397, O75405, Q17RW8, Q5BKS9, Q5QGN2, Q69YI5, Q6IN64, Q6ZN53, Q6ZS25, Q8N3R0, Q8TCN0, Q9C0D1, Q9H696, Q9NZW9 | Gene names | SETD2, HIF1, HYPB, KIAA1732, SET2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36-specific (EC 2.1.1.43) (SET domain-containing protein 2) (hSET2) (Huntingtin- interacting protein HYPB) (Huntingtin-interacting protein 1) (HIF-1) (p231HBP). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.469558 (rank : 9) | θ value | 2.21117e-06 (rank : 20) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH1_MOUSE
|
||||||
| NC score | 0.469129 (rank : 10) | θ value | 2.21117e-06 (rank : 21) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P70351, Q9R089 | Gene names | Ezh1, Enx2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
EZH2_HUMAN
|
||||||
| NC score | 0.464149 (rank : 11) | θ value | 1.09739e-05 (rank : 22) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q15910, Q15755, Q92857 | Gene names | EZH2 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.458273 (rank : 12) | θ value | 1.99992e-07 (rank : 19) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
EZH2_MOUSE
|
||||||
| NC score | 0.456659 (rank : 13) | θ value | 5.44631e-05 (rank : 23) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61188, Q9R090 | Gene names | Ezh2, Enx1h | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 2 (ENX-1). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.428437 (rank : 14) | θ value | 3.89403e-11 (rank : 11) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.426938 (rank : 15) | θ value | 3.08544e-08 (rank : 16) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.424029 (rank : 16) | θ value | 3.08544e-08 (rank : 17) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.423731 (rank : 17) | θ value | 9.59137e-10 (rank : 13) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.356773 (rank : 18) | θ value | 1.05554e-24 (rank : 4) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT2_MOUSE
|
||||||
| NC score | 0.353924 (rank : 19) | θ value | 4.01107e-24 (rank : 5) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Z148, Q6PE08, Q8K4R6, Q8K4R7, Q9Z149 | Gene names | Ehmt2, Bat8, G9a, Ng36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.348462 (rank : 20) | θ value | 1.16704e-23 (rank : 6) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.333410 (rank : 21) | θ value | 1.63604e-09 (rank : 14) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.323580 (rank : 22) | θ value | 6.87365e-08 (rank : 18) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.311801 (rank : 23) | θ value | 3.08544e-08 (rank : 15) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SETD8_MOUSE
|
||||||
| NC score | 0.310906 (rank : 24) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2YDW7, Q8C0J9 | Gene names | SETD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8). | |||||
|
SETD8_HUMAN
|
||||||
| NC score | 0.308985 (rank : 25) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9NQR1, Q86W83, Q8TD09 | Gene names | SETD8, PRSET7, SET07, SET8 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H4 lysine-20 specific (EC 2.1.1.43) (Histone H4-K20 methyltransferase) (H4-K20-HMTase) (SET domain-containing lysine methyltransferase 8) (SET domain-containing protein 8) (PR/SET domain-containing protein 07) (PR/SET07) (PR-Set7). | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.084712 (rank : 26) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.077614 (rank : 27) | θ value | 0.21417 (rank : 32) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.077205 (rank : 28) | θ value | 0.0736092 (rank : 27) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.074397 (rank : 29) | θ value | 0.00509761 (rank : 24) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
HN1_MOUSE
|
||||||
| NC score | 0.071729 (rank : 30) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97825 | Gene names | Hn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hematological and neurological expressed 1 protein. | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.066741 (rank : 31) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.065561 (rank : 32) | θ value | 3.0926 (rank : 76) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.065174 (rank : 33) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.065120 (rank : 34) | θ value | 0.00665767 (rank : 25) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.064540 (rank : 35) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.064470 (rank : 36) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.063541 (rank : 37) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.063541 (rank : 38) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.063004 (rank : 39) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.062863 (rank : 40) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.062772 (rank : 41) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.062551 (rank : 42) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.062082 (rank : 43) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.061777 (rank : 44) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.061462 (rank : 45) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.061377 (rank : 46) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.060845 (rank : 47) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
VPS72_MOUSE
|
||||||
| NC score | 0.060362 (rank : 48) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62481, Q3U2N4, Q810A9, Q99K81 | Gene names | Vps72, Tcfl1, Yl1 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 72 homolog (Transcription factor-like 1) (Protein YL-1). | |||||
|
SETD7_MOUSE
|
||||||
| NC score | 0.058870 (rank : 49) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VHL1 | Gene names | Setd7, Set7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7). | |||||
|
SETD7_HUMAN
|
||||||
| NC score | 0.057832 (rank : 50) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WTS6, Q9C0E6 | Gene names | SETD7, KIAA1717, SET7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET7 (EC 2.1.1.43) (Histone H3-K4 methyltransferase) (H3-K4-HMTase) (SET domain-containing protein 7) (Set9) (SET7/9). | |||||
|
F126A_MOUSE
|
||||||
| NC score | 0.057787 (rank : 51) | θ value | 0.163984 (rank : 30) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6P9N1, Q99NC4 | Gene names | Fam126a, Drctnnb1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM126A (Down-regulated by CTNNB1 protein A). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.057768 (rank : 52) | θ value | 1.06291 (rank : 49) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.056976 (rank : 53) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.054793 (rank : 54) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
AKAP2_MOUSE
|
||||||
| NC score | 0.054119 (rank : 55) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O54931, O54932, O54933 | Gene names | Akap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2) (AKAP expressed in kidney and lung) (AKAP-KL). | |||||
|
TARSH_HUMAN
|
||||||
| NC score | 0.053273 (rank : 56) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 768 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7Z7G0, Q6ZW20, Q6ZW22, Q9C082, Q9UFI6 | Gene names | ABI3BP, NESHBP, TARSH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Target of Nesh-SH3 precursor (Tarsh) (Nesh-binding protein) (NeshBP) (ABI gene family member 3-binding protein). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.051985 (rank : 57) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
ANR22_HUMAN
|
||||||
| NC score | 0.051178 (rank : 58) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5VYY1, Q8WU06 | Gene names | ANKRD22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 22. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.050838 (rank : 59) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.050819 (rank : 60) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.050644 (rank : 61) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.050637 (rank : 62) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.050582 (rank : 63) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.050501 (rank : 64) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.048276 (rank : 65) | θ value | 3.0926 (rank : 77) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.047541 (rank : 66) | θ value | 4.03905 (rank : 86) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.046410 (rank : 67) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.046073 (rank : 68) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.044999 (rank : 69) | θ value | 6.88961 (rank : 121) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.044956 (rank : 70) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
F126A_HUMAN
|
||||||
| NC score | 0.044593 (rank : 71) | θ value | 2.36792 (rank : 69) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BYI3, Q6N010, Q75MR4, Q7LDZ4, Q96MX1, Q96NQ6 | Gene names | FAM126A, DRCTNNB1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM126A (Down-regulated by CTNNB1 protein A). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.043659 (rank : 72) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.042521 (rank : 73) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MYCD_HUMAN
|
||||||
| NC score | 0.041987 (rank : 74) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8IZQ8, Q8N7Q1 | Gene names | MYOCD, MYCD | |||
|
Domain Architecture |

|
|||||
| Description | Myocardin. | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.041685 (rank : 75) | θ value | 0.163984 (rank : 31) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
K1543_MOUSE
|
||||||
| NC score | 0.041683 (rank : 76) | θ value | 4.03905 (rank : 87) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q80VC9, Q5DTW9, Q8BUZ0 | Gene names | Kiaa1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
CT055_MOUSE
|
||||||
| NC score | 0.041202 (rank : 77) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8R184, Q8BQE5, Q8K024, Q8VEM7, Q9CU80 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55 homolog. | |||||
|
RASF8_MOUSE
|
||||||
| NC score | 0.038936 (rank : 78) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CJ96, Q8CCW4, Q9CU91 | Gene names | Rassf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras association domain-containing protein 8 (Carcinoma-associated protein HOJ-1 homolog). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.038329 (rank : 79) | θ value | 4.03905 (rank : 93) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.037485 (rank : 80) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.037095 (rank : 81) | θ value | 2.36792 (rank : 70) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
CT2NL_MOUSE
|
||||||
| NC score | 0.036909 (rank : 82) | θ value | 1.81305 (rank : 60) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 902 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99LJ0, Q6ZPR0, Q8BSV1 | Gene names | Cttnbp2nl, Kiaa1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.035290 (rank : 83) | θ value | 3.0926 (rank : 80) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
WDR7_HUMAN
|
||||||
| NC score | 0.034579 (rank : 84) | θ value | 0.0736092 (rank : 28) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E6, Q86UX5, Q86VP2, Q96PS7 | Gene names | WDR7, KIAA0541, TRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 7 (TGF-beta resistance-associated protein TRAG) (Rabconnectin-3 beta). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.033259 (rank : 85) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.033216 (rank : 86) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.032518 (rank : 87) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.031281 (rank : 88) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.030530 (rank : 89) | θ value | 4.03905 (rank : 90) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.030156 (rank : 90) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
TACC2_MOUSE
|
||||||
| NC score | 0.029667 (rank : 91) | θ value | 3.0926 (rank : 82) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 774 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9JJG0 | Gene names | Tacc2 | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 2. | |||||
|
ZF106_HUMAN
|
||||||
| NC score | 0.029362 (rank : 92) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H2Y7, Q6NSD9, Q6PEK1, Q86T43, Q86T45, Q86T50, Q86T58, Q86TA9, Q96M37, Q9H7B8 | Gene names | ZFP106, ZNF474 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 106 homolog (Zfp-106) (Zinc finger protein 474). | |||||
|
BLNK_MOUSE
|
||||||
| NC score | 0.029350 (rank : 93) | θ value | 4.03905 (rank : 83) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9QUN3, O88504 | Gene names | Blnk, Bash, Ly57, Slp65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell linker protein (Cytoplasmic adapter protein) (B-cell adapter containing SH2 domain protein) (B-cell adapter containing Src homology 2 domain protein) (Src homology 2 domain-containing leukocyte protein of 65 kDa) (Slp-65) (Lymphocyte antigen 57). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.029266 (rank : 94) | θ value | 5.27518 (rank : 102) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
RN165_HUMAN
|
||||||
| NC score | 0.029248 (rank : 95) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.028679 (rank : 96) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
EP15R_MOUSE
|
||||||
| NC score | 0.028432 (rank : 97) | θ value | 2.36792 (rank : 68) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q60902, Q8CB60, Q8CB70, Q91WH8 | Gene names | Eps15l1, Eps15-rs, Eps15R | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15-like 1 (Eps15-related protein) (Eps15R) (Epidermal growth factor receptor pathway substrate 15-related sequence) (Eps15-rs). | |||||
|
WTAP_HUMAN
|
||||||
| NC score | 0.028265 (rank : 98) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q15007, Q96T28, Q9BYJ7, Q9H4E2 | Gene names | WTAP, KIAA0105 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wilms' tumor 1-associating protein (WT1-associated protein) (Putative pre-mRNA-splicing regulator female-lethal(2D) homolog). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.028023 (rank : 99) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.027849 (rank : 100) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
CHD2_HUMAN
|
||||||
| NC score | 0.027651 (rank : 101) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14647 | Gene names | CHD2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 2 (EC 3.6.1.-) (ATP- dependent helicase CHD2) (CHD-2). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.027648 (rank : 102) | θ value | 8.99809 (rank : 133) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.027269 (rank : 103) | θ value | 4.03905 (rank : 84) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.027178 (rank : 104) | θ value | 8.99809 (rank : 136) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.026780 (rank : 105) | θ value | 8.99809 (rank : 137) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
TSC1_HUMAN
|
||||||
| NC score | 0.026774 (rank : 106) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 765 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q92574 | Gene names | TSC1, KIAA0243, TSC | |||
|
Domain Architecture |

|
|||||
| Description | Hamartin (Tuberous sclerosis 1 protein). | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.026477 (rank : 107) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.026271 (rank : 108) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.026197 (rank : 109) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
EXTL1_MOUSE
|
||||||
| NC score | 0.025411 (rank : 110) | θ value | 0.365318 (rank : 36) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JKV7 | Gene names | Extl1 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 1 (EC 2.4.1.224) (Glucuronosyl-N-acetylglucosaminyl- proteoglycan 4-alpha-N-acetylglucosaminyltransferase) (Exostosin-L) (Multiple exostosis-like protein). | |||||
|
FIBA_HUMAN
|
||||||
| NC score | 0.025367 (rank : 111) | θ value | 0.0961366 (rank : 29) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02671, Q9BX62, Q9UCH2 | Gene names | FGA | |||
|
Domain Architecture |

|
|||||
| Description | Fibrinogen alpha chain precursor [Contains: Fibrinopeptide A]. | |||||
|
SELS_HUMAN
|
||||||
| NC score | 0.025346 (rank : 112) | θ value | 4.03905 (rank : 91) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQE4, Q3B771, Q9P0I6 | Gene names | SELS, VIMP | |||
|
Domain Architecture |

|
|||||
| Description | Selenoprotein S (VCP-interacting membrane protein). | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.024646 (rank : 113) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.024345 (rank : 114) | θ value | 5.27518 (rank : 99) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.024088 (rank : 115) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
FLNB_HUMAN
|
||||||
| NC score | 0.023677 (rank : 116) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75369, Q13706, Q8WXS9, Q8WXT0, Q8WXT1, Q8WXT2, Q9NRB5, Q9NT26, Q9UEV9 | Gene names | FLNB, FLN1L, FLN3, TABP, TAP | |||
|
Domain Architecture |

|
|||||
| Description | Filamin-B (FLN-B) (Beta-filamin) (Actin-binding-like protein) (Thyroid autoantigen) (Truncated actin-binding protein) (Truncated ABP) (ABP- 280 homolog) (ABP-278) (Filamin 3) (Filamin homolog 1) (Fh1). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.022926 (rank : 117) | θ value | 0.279714 (rank : 34) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.022894 (rank : 118) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.022771 (rank : 119) | θ value | 8.99809 (rank : 128) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.022308 (rank : 120) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.021516 (rank : 121) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.021097 (rank : 122) | θ value | 5.27518 (rank : 103) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.020631 (rank : 123) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
UT14A_HUMAN
|
||||||
| NC score | 0.019985 (rank : 124) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BVJ6, Q5JYF1 | Gene names | UTP14A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar RNA-associated protein 14 homolog A (Antigen NY-CO- 16). | |||||
|
CENPJ_HUMAN
|
||||||
| NC score | 0.018998 (rank : 125) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC77, Q5T6R5, Q96KS5, Q9C067 | Gene names | CENPJ, CPAP, LAP, LIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein J (CENP-J) (Centrosomal P4.1-associated protein) (LAG-3-associated protein) (LYST-interacting protein 1). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.018794 (rank : 126) | θ value | 6.88961 (rank : 120) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.018767 (rank : 127) | θ value | 6.88961 (rank : 119) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.017859 (rank : 128) | θ value | 8.99809 (rank : 130) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
CNGB3_HUMAN
|
||||||
| NC score | 0.016958 (rank : 129) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NQW8, Q9NRE9 | Gene names | CNGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic nucleotide-gated cation channel beta 3 (CNG channel beta 3) (Cyclic nucleotide-gated channel beta 3) (Cone photoreceptor cGMP- gated channel subunit beta) (Cyclic nucleotide-gated cation channel modulatory subunit). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.016009 (rank : 130) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
HS74L_HUMAN
|
||||||
| NC score | 0.015684 (rank : 131) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O95757, Q4W5M5, Q8IWA2 | Gene names | HSPA4L, APG1, OSP94 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock 70 kDa protein 4L (Osmotic stress protein 94) (Heat shock 70-related protein APG-1). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.015576 (rank : 132) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
NTNG1_HUMAN
|
||||||
| NC score | 0.015379 (rank : 133) | θ value | 3.0926 (rank : 78) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NCOA7_MOUSE
|
||||||
| NC score | 0.014936 (rank : 134) | θ value | 4.03905 (rank : 88) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6DFV7, Q3TAR1, Q3UNB3, Q66K05, Q6ZQM4, Q8BJ15, Q8BJ39 | Gene names | Ncoa7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 7. | |||||
|
PALB2_MOUSE
|
||||||
| NC score | 0.014749 (rank : 135) | θ value | 6.88961 (rank : 117) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U0P1, Q6NZG9, Q7TMQ4, Q8CEA9 | Gene names | Palb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Partner and localizer of BRCA2. | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.014487 (rank : 136) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
MRE11_HUMAN
|
||||||
| NC score | 0.013451 (rank : 137) | θ value | 6.88961 (rank : 116) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P49959, O43475 | Gene names | MRE11A, HNGS1, MRE11 | |||
|
Domain Architecture |

|
|||||
| Description | Double-strand break repair protein MRE11A (MRE11 homolog 1) (MRE11 meiotic recombination 11 homolog A). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.012821 (rank : 138) | θ value | 3.0926 (rank : 81) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.012674 (rank : 139) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
CACB1_HUMAN
|
||||||
| NC score | 0.012399 (rank : 140) | θ value | 5.27518 (rank : 96) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02641, O15331, Q02639, Q02640, Q9C085 | Gene names | CACNB1, CACNLB1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
CACB1_MOUSE
|
||||||
| NC score | 0.012349 (rank : 141) | θ value | 5.27518 (rank : 97) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R3Z5, O88517, Q7TPF2, Q8R3Z6, Q9EPT9 | Gene names | Cacnb1, Cacnlb1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-1 (CAB1) (Calcium channel voltage-dependent subunit beta 1). | |||||
|
RNF17_HUMAN
|
||||||
| NC score | 0.012245 (rank : 142) | θ value | 8.99809 (rank : 135) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXT8, Q9BXT7 | Gene names | RNF17 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 17. | |||||
|
SSH3_HUMAN
|
||||||
| NC score | 0.011887 (rank : 143) | θ value | 5.27518 (rank : 104) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TE77, Q6PK42, Q76I75, Q8N9L8, Q8WYL0, Q9NV45, Q9NWZ7 | Gene names | SSH3, SSH3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein phosphatase Slingshot homolog 3 (EC 3.1.3.48) (EC 3.1.3.16) (SSH-3L) (hSSH-3L). | |||||
|
DDX58_MOUSE
|
||||||
| NC score | 0.011772 (rank : 144) | θ value | 4.03905 (rank : 85) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6Q899, Q8C320, Q8C5I3, Q8C7T2 | Gene names | Ddx58 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX58 (EC 3.6.1.-) (DEAD-box protein 58) (Retinoic acid-inducible gene 1 protein) (RIG-1) (RIG-I). | |||||
|
FOXD1_MOUSE
|
||||||
| NC score | 0.011670 (rank : 145) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
PITM1_HUMAN
|
||||||
| NC score | 0.011094 (rank : 146) | θ value | 4.03905 (rank : 89) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00562, Q6T7X3, Q8TBN3, Q9BZ73 | Gene names | PITPNM1, DRES9, NIR2, PITPNM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog). | |||||
|
ETV4_HUMAN
|
||||||
| NC score | 0.011065 (rank : 147) | θ value | 1.81305 (rank : 61) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P43268, Q96AW9 | Gene names | ETV4, E1AF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 4 (Adenovirus E1A enhancer-binding protein) (E1A-F). | |||||
|
PDIP3_MOUSE
|
||||||
| NC score | 0.011016 (rank : 148) | θ value | 8.99809 (rank : 132) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BG81, Q6PE83, Q80X99, Q8CI28 | Gene names | Poldip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polymerase delta-interacting protein 3. | |||||
|
NTNG1_MOUSE
|
||||||
| NC score | 0.010918 (rank : 149) | θ value | 8.99809 (rank : 131) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
KRA11_HUMAN
|
||||||
| NC score | 0.010615 (rank : 150) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07627, Q96S60, Q96S67 | Gene names | KRTAP1-1, B2A, KAP1.1, KAP1.6, KAP1.7, KRTAP1.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 1-1 (Keratin-associated protein 1.1) (High sulfur keratin-associated protein 1.1) (Keratin-associated protein 1.6) (Keratin-associated protein 1.7). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.010368 (rank : 151) | θ value | 5.27518 (rank : 100) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.009678 (rank : 152) | θ value | 5.27518 (rank : 101) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
VMDL3_HUMAN
|
||||||
| NC score | 0.009082 (rank : 153) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N1M1, Q8N356, Q8NFT9, Q9BR80 | Gene names | VMD2L3 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-4 (Vitelliform macular dystrophy 2-like protein 3). | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.008836 (rank : 154) | θ value | 8.99809 (rank : 129) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
FOXO1_HUMAN
|
||||||
| NC score | 0.008580 (rank : 155) | θ value | 5.27518 (rank : 98) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12778, O43523, Q5VYC7, Q6NSK6 | Gene names | FOXO1A, FKHR | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein O1A (Forkhead in rhabdomyosarcoma). | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.007669 (rank : 156) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
LRCH3_MOUSE
|
||||||
| NC score | 0.006578 (rank : 157) | θ value | 3.0926 (rank : 75) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
SEM6B_MOUSE
|
||||||
| NC score | 0.006075 (rank : 158) | θ value | 4.03905 (rank : 92) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54951 | Gene names | Sema6b, Seman | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6B precursor (Semaphorin VIB) (Sema VIB) (Semaphorin N) (Sema N). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.006058 (rank : 159) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
CD2L7_HUMAN
|
||||||
| NC score | 0.005506 (rank : 160) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NYV4, O94978 | Gene names | CRKRS, CRK7, KIAA0904 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle 2-related protein kinase 7 (EC 2.7.11.22) (CDC2- related protein kinase 7) (Cdc2-related kinase, arginine/serine-rich) (CrkRS). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.005102 (rank : 161) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
POK3_HUMAN
|
||||||
| NC score | 0.004817 (rank : 162) | θ value | 6.88961 (rank : 118) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WJR5, O15312 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Pol protein (HERV-K(C19) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
PZRN4_HUMAN
|
||||||
| NC score | 0.004150 (rank : 163) | θ value | 8.99809 (rank : 134) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZMN7, Q6N052, Q8IUU1, Q9NTP7 | Gene names | PDZRN4, LNX4, SEMCAP3L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 4 (Ligand of Numb-protein X 4) (SEMACAP3-like protein). | |||||
|
TMPS2_MOUSE
|
||||||
| NC score | 0.004050 (rank : 164) | θ value | 4.03905 (rank : 94) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TRI62_MOUSE
|
||||||
| NC score | 0.003985 (rank : 165) | θ value | 8.99809 (rank : 138) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 478 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80V85 | Gene names | Trim62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 62. | |||||
|
OR2J1_HUMAN
|
||||||
| NC score | 0.002424 (rank : 166) | θ value | 0.47712 (rank : 41) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 758 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZK6, Q9GZK1 | Gene names | OR2J1 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 2J1 (Olfactory receptor 6-5) (OR6-5) (Hs6M1-4). | |||||
|
OR2J3_HUMAN
|
||||||
| NC score | 0.000259 (rank : 167) | θ value | 3.0926 (rank : 79) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O76001, Q96R15, Q9GZK5, Q9GZL4, Q9GZL5 | Gene names | OR2J3 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 2J3 (Olfactory receptor 6-6) (OR6-6) (Hs6M1-3). | |||||
|
ZIC3_MOUSE
|
||||||
| NC score | -0.000880 (rank : 168) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62521 | Gene names | Zic3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein ZIC 3 (Zinc finger protein of the cerebellum 3). | |||||
|
ZIC3_HUMAN
|
||||||
| NC score | -0.000886 (rank : 169) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O60481 | Gene names | ZIC3 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein ZIC 3 (Zinc finger protein of the cerebellum 3). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | -0.001216 (rank : 170) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
ZN445_HUMAN
|
||||||
| NC score | -0.002917 (rank : 171) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 141 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59923 | Gene names | ZNF445, ZNF168 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 445. | |||||