Please be patient as the page loads
|
ECHP_MOUSE
|
||||||
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ECHP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985805 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
ECHP_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
ECHA_HUMAN
|
||||||
| θ value | 7.45998e-71 (rank : 3) | NC score | 0.909881 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
HCDH_HUMAN
|
||||||
| θ value | 2.34606e-32 (rank : 4) | NC score | 0.706090 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |
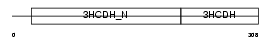
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
HCDH_MOUSE
|
||||||
| θ value | 1.98606e-31 (rank : 5) | NC score | 0.701687 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |
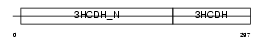
|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
ECHM_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 6) | NC score | 0.722487 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| θ value | 2.20094e-22 (rank : 7) | NC score | 0.712991 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
CRYL1_HUMAN
|
||||||
| θ value | 7.58209e-15 (rank : 8) | NC score | 0.526757 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2S2, Q7Z4Z9 | Gene names | CRYL1, CRY | |||
|
Domain Architecture |
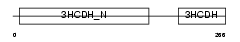
|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
AUHM_MOUSE
|
||||||
| θ value | 2.88119e-14 (rank : 9) | NC score | 0.673577 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 10) | NC score | 0.668239 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
CRYL1_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 11) | NC score | 0.469491 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KP3, Q542R9, Q8R4W7 | Gene names | Cryl1, Cry | |||
|
Domain Architecture |
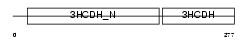
|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
ECH1_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 12) | NC score | 0.618540 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
D3D2_MOUSE
|
||||||
| θ value | 4.59992e-12 (rank : 13) | NC score | 0.548295 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42125 | Gene names | Dci | |||
|
Domain Architecture |
|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CDYL1_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 14) | NC score | 0.444689 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 15) | NC score | 0.447059 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
D3D2_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 16) | NC score | 0.542522 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42126, Q13290, Q7Z2L6, Q9BUB8, Q9BW05 | Gene names | DCI | |||
|
Domain Architecture |
|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CDYL2_HUMAN
|
||||||
| θ value | 1.63604e-09 (rank : 17) | NC score | 0.419464 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
PECI_MOUSE
|
||||||
| θ value | 2.79066e-09 (rank : 18) | NC score | 0.444048 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CDY1_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 19) | NC score | 0.442549 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| θ value | 1.06045e-08 (rank : 20) | NC score | 0.439709 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
ECH1_MOUSE
|
||||||
| θ value | 2.36244e-08 (rank : 21) | NC score | 0.596468 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
FAK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 22) | NC score | 0.000908 (rank : 34) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.011439 (rank : 31) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
3HIDH_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.017608 (rank : 30) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31937, Q9UDN3 | Gene names | HIBADH | |||
|
Domain Architecture |
|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.007108 (rank : 32) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MOGT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.006075 (rank : 33) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZV4, Q9DCL0 | Gene names | Mogat1, Dgat2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2-acylglycerol O-acyltransferase 1 (EC 2.3.1.22) (Monoacylglycerol O- acyltransferase 1) (Acyl CoA:monoacylglycerol acyltransferase 1) (MGAT1) (Diacylglycerol acyltransferase 2-like protein 1). | |||||
|
CBX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.057947 (rank : 27) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.057947 (rank : 28) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.058112 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.062466 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.058027 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.058769 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.053124 (rank : 29) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
PECI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.326245 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
ECHP_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9DBM2 | Gene names | Ehhadh | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
ECHP_HUMAN
|
||||||
| NC score | 0.985805 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q08426 | Gene names | EHHADH, ECHD | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal bifunctional enzyme (PBE) (PBFE) [Includes: Enoyl-CoA hydratase (EC 4.2.1.17); 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8); 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35)]. | |||||
|
ECHA_HUMAN
|
||||||
| NC score | 0.909881 (rank : 3) | θ value | 7.45998e-71 (rank : 3) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P40939, Q16679, Q96GT7 | Gene names | HADHA, HADH | |||
|
Domain Architecture |

|
|||||
| Description | Trifunctional enzyme subunit alpha, mitochondrial precursor (TP-alpha) (78 kDa gastrin-binding protein) [Includes: Long-chain enoyl-CoA hydratase (EC 4.2.1.17); Long chain 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.211)]. | |||||
|
ECHM_MOUSE
|
||||||
| NC score | 0.722487 (rank : 4) | θ value | 2.12685e-25 (rank : 6) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BH95, Q3TJK2, Q6GQS2, Q6PEN1, Q99LX7 | Gene names | Echs1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
ECHM_HUMAN
|
||||||
| NC score | 0.712991 (rank : 5) | θ value | 2.20094e-22 (rank : 7) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P30084, O00739, Q5VWY1, Q96H54 | Gene names | ECHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) (Short chain enoyl-CoA hydratase) (SCEH) (Enoyl-CoA hydratase 1). | |||||
|
HCDH_HUMAN
|
||||||
| NC score | 0.706090 (rank : 6) | θ value | 2.34606e-32 (rank : 4) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16836, O00324, O00397, O00753 | Gene names | HADHSC, HAD, SCHAD | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
HCDH_MOUSE
|
||||||
| NC score | 0.701687 (rank : 7) | θ value | 1.98606e-31 (rank : 5) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61425, Q3TF75, Q3THK8, Q3UFI0, Q8K149, Q925U9 | Gene names | Hadhsc, Hadh, Mschad, Schad | |||
|
Domain Architecture |

|
|||||
| Description | Short chain 3-hydroxyacyl-CoA dehydrogenase, mitochondrial precursor (EC 1.1.1.35) (HCDH) (Medium and short chain L-3-hydroxyacyl-coenzyme A dehydrogenase). | |||||
|
AUHM_MOUSE
|
||||||
| NC score | 0.673577 (rank : 8) | θ value | 2.88119e-14 (rank : 9) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JLZ3, Q80YD7, Q8QZS0, Q9CY78, Q9D155 | Gene names | Auh | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding enoyl-CoA hydratase) (muAUH). | |||||
|
AUMH_HUMAN
|
||||||
| NC score | 0.668239 (rank : 9) | θ value | 8.38298e-14 (rank : 10) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13825, Q8WUE4 | Gene names | AUH | |||
|
Domain Architecture |

|
|||||
| Description | Methylglutaconyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.18) (AU-specific RNA-binding enoyl-CoA hydratase) (AU-binding protein/enoyl-CoA hydratase). | |||||
|
ECH1_HUMAN
|
||||||
| NC score | 0.618540 (rank : 10) | θ value | 2.43908e-13 (rank : 12) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13011, Q8WVX0, Q96EZ9 | Gene names | ECH1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
ECH1_MOUSE
|
||||||
| NC score | 0.596468 (rank : 11) | θ value | 2.36244e-08 (rank : 21) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O35459, Q5M8P6 | Gene names | Ech1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta3,5-delta2,4-dienoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.-). | |||||
|
D3D2_MOUSE
|
||||||
| NC score | 0.548295 (rank : 12) | θ value | 4.59992e-12 (rank : 13) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42125 | Gene names | Dci | |||
|
Domain Architecture |

|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
D3D2_HUMAN
|
||||||
| NC score | 0.542522 (rank : 13) | θ value | 1.25267e-09 (rank : 16) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P42126, Q13290, Q7Z2L6, Q9BUB8, Q9BW05 | Gene names | DCI | |||
|
Domain Architecture |

|
|||||
| Description | 3,2-trans-enoyl-CoA isomerase, mitochondrial precursor (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CRYL1_HUMAN
|
||||||
| NC score | 0.526757 (rank : 14) | θ value | 7.58209e-15 (rank : 8) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2S2, Q7Z4Z9 | Gene names | CRYL1, CRY | |||
|
Domain Architecture |

|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
CRYL1_MOUSE
|
||||||
| NC score | 0.469491 (rank : 15) | θ value | 2.43908e-13 (rank : 11) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99KP3, Q542R9, Q8R4W7 | Gene names | Cryl1, Cry | |||
|
Domain Architecture |

|
|||||
| Description | Lambda-crystallin homolog. | |||||
|
CDYL_MOUSE
|
||||||
| NC score | 0.447059 (rank : 16) | θ value | 9.59137e-10 (rank : 15) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTK2 | Gene names | Cdyl | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
CDYL1_HUMAN
|
||||||
| NC score | 0.444689 (rank : 17) | θ value | 5.62301e-10 (rank : 14) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y232 | Gene names | CDYL, CDYL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein (CDY-like). | |||||
|
PECI_MOUSE
|
||||||
| NC score | 0.444048 (rank : 18) | θ value | 2.79066e-09 (rank : 18) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WUR2 | Gene names | Peci | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase). | |||||
|
CDY1_HUMAN
|
||||||
| NC score | 0.442549 (rank : 19) | θ value | 6.21693e-09 (rank : 19) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6F8, O14600 | Gene names | CDY1 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 1. | |||||
|
CDY2_HUMAN
|
||||||
| NC score | 0.439709 (rank : 20) | θ value | 1.06045e-08 (rank : 20) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y6F7 | Gene names | CDY2A, CDY2 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific chromodomain protein Y 2. | |||||
|
CDYL2_HUMAN
|
||||||
| NC score | 0.419464 (rank : 21) | θ value | 1.63604e-09 (rank : 17) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8N8U2 | Gene names | CDYL2 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain Y-like protein 2 (CDY-like 2). | |||||
|
PECI_HUMAN
|
||||||
| NC score | 0.326245 (rank : 22) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75521, Q5JYK5, Q8N0X0, Q9BUE9, Q9H0T9, Q9NQH1, Q9NYH7, Q9UN55 | Gene names | PECI, DRS1, HCA88 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal 3,2-trans-enoyl-CoA isomerase (EC 5.3.3.8) (Dodecenoyl-CoA isomerase) (Delta(3),delta(2)-enoyl-CoA isomerase) (D3,D2-enoyl-CoA isomerase) (DBI-related protein 1) (DRS-1) (Hepatocellular carcinoma- associated antigen 88) (NY-REN-1 antigen). | |||||
|
CBX3_MOUSE
|
||||||
| NC score | 0.062466 (rank : 23) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P23198, Q921C4 | Gene names | Cbx3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (M32). | |||||
|
CBX5_MOUSE
|
||||||
| NC score | 0.058769 (rank : 24) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61686, Q9CS35, Q9CXD1 | Gene names | Cbx5, Hp1a | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha). | |||||
|
CBX3_HUMAN
|
||||||
| NC score | 0.058112 (rank : 25) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13185, Q96CD7, Q99409, Q9BVS3, Q9P0Z6 | Gene names | CBX3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 3 (Heterochromatin protein 1 homolog gamma) (HP1 gamma) (Modifier 2 protein) (HECH). | |||||
|
CBX5_HUMAN
|
||||||
| NC score | 0.058027 (rank : 26) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P45973 | Gene names | CBX5, HP1A | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 5 (Heterochromatin protein 1 homolog alpha) (HP1 alpha) (Antigen p25). | |||||
|
CBX1_HUMAN
|
||||||
| NC score | 0.057947 (rank : 27) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P83916, P23197 | Gene names | CBX1, CBX | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25) (HP1Hsbeta) (p25beta). | |||||
|
CBX1_MOUSE
|
||||||
| NC score | 0.057947 (rank : 28) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P83917, P23197 | Gene names | Cbx1, Cbx | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 1 (Heterochromatin protein 1 homolog beta) (HP1 beta) (Modifier 1 protein) (M31) (Heterochromatin protein p25). | |||||
|
CBX7_MOUSE
|
||||||
| NC score | 0.053124 (rank : 29) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VDS3 | Gene names | Cbx7, D15Ertd417e | |||
|
Domain Architecture |

|
|||||
| Description | Chromobox protein homolog 7. | |||||
|
3HIDH_HUMAN
|
||||||
| NC score | 0.017608 (rank : 30) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31937, Q9UDN3 | Gene names | HIBADH | |||
|
Domain Architecture |

|
|||||
| Description | 3-hydroxyisobutyrate dehydrogenase, mitochondrial precursor (EC 1.1.1.31) (HIBADH). | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.011439 (rank : 31) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.007108 (rank : 32) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MOGT1_MOUSE
|
||||||
| NC score | 0.006075 (rank : 33) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91ZV4, Q9DCL0 | Gene names | Mogat1, Dgat2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2-acylglycerol O-acyltransferase 1 (EC 2.3.1.22) (Monoacylglycerol O- acyltransferase 1) (Acyl CoA:monoacylglycerol acyltransferase 1) (MGAT1) (Diacylglycerol acyltransferase 2-like protein 1). | |||||
|
FAK2_HUMAN
|
||||||
| NC score | 0.000908 (rank : 34) | θ value | 1.81305 (rank : 22) | |||
| Query Neighborhood Hits | 26 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||