Please be patient as the page loads
|
TAIP2_MOUSE
|
||||||
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TAIP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996177 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAIP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAI12_HUMAN
|
||||||
| θ value | 3.1125e-93 (rank : 3) | NC score | 0.954691 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TAI12_MOUSE
|
||||||
| θ value | 4.49445e-92 (rank : 4) | NC score | 0.953255 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
AXUD1_HUMAN
|
||||||
| θ value | 5.53243e-66 (rank : 5) | NC score | 0.916170 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S65 | Gene names | AXUD1, TAIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3) (URAX1 protein). | |||||
|
AXUD1_MOUSE
|
||||||
| θ value | 1.96772e-63 (rank : 6) | NC score | 0.914079 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 7) | NC score | 0.019176 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.047406 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.053385 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.031379 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.044582 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.028411 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.014397 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.031503 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NCK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.014623 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |
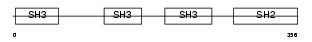
|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.022226 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.045954 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.026234 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.023742 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.015442 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.026157 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.013078 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
PZRN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.008550 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.003324 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
LRTM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.006609 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43300, Q7L770 | Gene names | LRRTM2, KIAA0416, LRRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 2 precursor (Leucine-rich repeat neuronal 2 protein). | |||||
|
WDR12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.008691 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJA4, Q9CST3, Q9JKF5 | Gene names | Wdr12 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 12 (YTM1 homolog). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.013464 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
FA13C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.013417 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.018340 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
N4BP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.019478 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
REV1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.025320 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.022842 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.019065 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.007946 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
EZH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.008069 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.019893 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TB182_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.014113 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.019035 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
MBTP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.010118 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
NALP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.007402 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.011978 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
WDR12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.006886 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZL7, Q96HU0, Q9NV80, Q9NYI8 | Gene names | WDR12 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 12 (YTM1 homolog). | |||||
|
TAIP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAIP2_HUMAN
|
||||||
| NC score | 0.996177 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAI12_HUMAN
|
||||||
| NC score | 0.954691 (rank : 3) | θ value | 3.1125e-93 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TAI12_MOUSE
|
||||||
| NC score | 0.953255 (rank : 4) | θ value | 4.49445e-92 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
AXUD1_HUMAN
|
||||||
| NC score | 0.916170 (rank : 5) | θ value | 5.53243e-66 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S65 | Gene names | AXUD1, TAIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3) (URAX1 protein). | |||||
|
AXUD1_MOUSE
|
||||||
| NC score | 0.914079 (rank : 6) | θ value | 1.96772e-63 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.053385 (rank : 7) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.047406 (rank : 8) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.045954 (rank : 9) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.044582 (rank : 10) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.031503 (rank : 11) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.031379 (rank : 12) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.028411 (rank : 13) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.026234 (rank : 14) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.026157 (rank : 15) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
REV1_HUMAN
|
||||||
| NC score | 0.025320 (rank : 16) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBZ9, O95941, Q9C0J4, Q9NUP2 | Gene names | REV1L, REV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein REV1 (EC 2.7.7.-) (Rev1-like terminal deoxycytidyl transferase) (Alpha integrin-binding protein 80) (AIBP80). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.023742 (rank : 17) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.022842 (rank : 18) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.022226 (rank : 19) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.019893 (rank : 20) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
N4BP2_HUMAN
|
||||||
| NC score | 0.019478 (rank : 21) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UW6, Q9NVK2, Q9P2D4 | Gene names | N4BP2, B3BP, KIAA1413 | |||
|
Domain Architecture |

|
|||||
| Description | NEDD4-binding protein 2 (EC 3.-.-.-) (N4BP2) (BCL-3-binding protein). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.019176 (rank : 22) | θ value | 0.21417 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.019065 (rank : 23) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.019035 (rank : 24) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.018340 (rank : 25) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.015442 (rank : 26) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
NCK2_HUMAN
|
||||||
| NC score | 0.014623 (rank : 27) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.014397 (rank : 28) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.014113 (rank : 29) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.013464 (rank : 30) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
FA13C_MOUSE
|
||||||
| NC score | 0.013417 (rank : 31) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBR2 | Gene names | Fam13c1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM13C1. | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.013078 (rank : 32) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.011978 (rank : 33) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
MBTP1_HUMAN
|
||||||
| NC score | 0.010118 (rank : 34) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14703, Q9UF67 | Gene names | MBTPS1, KIAA0091, S1P, SKI1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-bound transcription factor site 1 protease precursor (EC 3.4.21.-) (S1P endopeptidase) (Site-1 protease) (Subtilisin/kexin- isozyme 1) (SKI-1). | |||||
|
WDR12_MOUSE
|
||||||
| NC score | 0.008691 (rank : 35) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JJA4, Q9CST3, Q9JKF5 | Gene names | Wdr12 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 12 (YTM1 homolog). | |||||
|
PZRN3_HUMAN
|
||||||
| NC score | 0.008550 (rank : 36) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UPQ7, Q8N2N7, Q96CC2, Q9NSQ2 | Gene names | PDZRN3, KIAA1095, LNX3, SEMCAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ domain-containing RING finger protein 3 (Ligand of Numb-protein X 3) (Semaphorin cytoplasmic domain-associated protein 3) (SEMACAP3 protein). | |||||
|
EZH1_HUMAN
|
||||||
| NC score | 0.008069 (rank : 37) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92800, O43287, Q14459 | Gene names | EZH1, KIAA0388 | |||
|
Domain Architecture |

|
|||||
| Description | Enhancer of zeste homolog 1 (ENX-2). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.007946 (rank : 38) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
NALP5_HUMAN
|
||||||
| NC score | 0.007402 (rank : 39) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59047, Q86W29 | Gene names | NALP5, MATER | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Mater protein homolog). | |||||
|
WDR12_HUMAN
|
||||||
| NC score | 0.006886 (rank : 40) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9GZL7, Q96HU0, Q9NV80, Q9NYI8 | Gene names | WDR12 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 12 (YTM1 homolog). | |||||
|
LRTM2_HUMAN
|
||||||
| NC score | 0.006609 (rank : 41) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43300, Q7L770 | Gene names | LRRTM2, KIAA0416, LRRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 2 precursor (Leucine-rich repeat neuronal 2 protein). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.003324 (rank : 42) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||