Please be patient as the page loads
|
TAIP2_HUMAN
|
||||||
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TAIP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAIP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996177 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAI12_HUMAN
|
||||||
| θ value | 2.62879e-100 (rank : 3) | NC score | 0.958405 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TAI12_MOUSE
|
||||||
| θ value | 2.2254e-99 (rank : 4) | NC score | 0.956988 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
AXUD1_HUMAN
|
||||||
| θ value | 8.53533e-67 (rank : 5) | NC score | 0.917390 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S65 | Gene names | AXUD1, TAIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3) (URAX1 protein). | |||||
|
AXUD1_MOUSE
|
||||||
| θ value | 7.98882e-65 (rank : 6) | NC score | 0.915814 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 7) | NC score | 0.065639 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.057892 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
SREC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 9) | NC score | 0.025686 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.022033 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
CD226_HUMAN
|
||||||
| θ value | 1.38821 (rank : 11) | NC score | 0.029819 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15762 | Gene names | CD226, DNAM1 | |||
|
Domain Architecture |
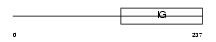
|
|||||
| Description | CD226 antigen precursor (DNAX accessory molecule 1) (DNAM-1). | |||||
|
FSTL4_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.016754 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.021095 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LATS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.005030 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.038742 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.002377 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.013436 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.036026 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CAC1E_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.009586 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
RRP44_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.036086 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
ZN638_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.016474 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
FSTL4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.013963 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.016278 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LRTM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.006586 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43300, Q7L770 | Gene names | LRRTM2, KIAA0416, LRRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 2 precursor (Leucine-rich repeat neuronal 2 protein). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.009982 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.027671 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
LYST_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.008602 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
NCK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.012285 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |
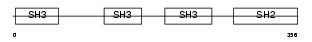
|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
DSRAD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.009892 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.019354 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.021458 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.006525 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.016846 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.011802 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
K0460_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.009043 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.019111 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.009004 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TAIP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WYN3 | Gene names | TAIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAIP2_MOUSE
|
||||||
| NC score | 0.996177 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P59055 | Gene names | Taip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 2 (TAIP-2). | |||||
|
TAI12_HUMAN
|
||||||
| NC score | 0.958405 (rank : 3) | θ value | 2.62879e-100 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H175 | Gene names | TAIP12, C12orf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
TAI12_MOUSE
|
||||||
| NC score | 0.956988 (rank : 4) | θ value | 2.2254e-99 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8BGQ2, Q3UIH1 | Gene names | Taip12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TGF-beta-induced apoptosis protein 12 (TAIP-12). | |||||
|
AXUD1_HUMAN
|
||||||
| NC score | 0.917390 (rank : 5) | θ value | 8.53533e-67 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96S65 | Gene names | AXUD1, TAIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3) (URAX1 protein). | |||||
|
AXUD1_MOUSE
|
||||||
| NC score | 0.915814 (rank : 6) | θ value | 7.98882e-65 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.065639 (rank : 7) | θ value | 0.0961366 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.057892 (rank : 8) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.038742 (rank : 9) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
RRP44_HUMAN
|
||||||
| NC score | 0.036086 (rank : 10) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y2L1, Q5W0P7, Q5W0P8, Q658Z7, Q7Z481, Q8WWI2, Q9UG36 | Gene names | DIS3, KIAA1008, RRP44 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome complex exonuclease RRP44 (EC 3.1.13.-) (Ribosomal RNA- processing protein 44) (DIS3 protein homolog). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.036026 (rank : 11) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
CD226_HUMAN
|
||||||
| NC score | 0.029819 (rank : 12) | θ value | 1.38821 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15762 | Gene names | CD226, DNAM1 | |||
|
Domain Architecture |

|
|||||
| Description | CD226 antigen precursor (DNAX accessory molecule 1) (DNAM-1). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.027671 (rank : 13) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.025686 (rank : 14) | θ value | 1.06291 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.022033 (rank : 15) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.021458 (rank : 16) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.021095 (rank : 17) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.019354 (rank : 18) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.019111 (rank : 19) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.016846 (rank : 20) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
FSTL4_MOUSE
|
||||||
| NC score | 0.016754 (rank : 21) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5STE3, Q5DU03 | Gene names | Fstl4, Kiaa1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4) (m- D/Bsp120I 1-1). | |||||
|
ZN638_HUMAN
|
||||||
| NC score | 0.016474 (rank : 22) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14966, Q53R34, Q5XJ05, Q68DP3, Q6P2H2, Q7Z3T7, Q8NF92, Q8TCA1, Q9H2G1, Q9NP37 | Gene names | ZNF638, NP220, ZFML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 638 (Nuclear protein 220) (Zinc-finger matrin-like protein) (Cutaneous T-cell lymphoma-associated antigen se33-1) (CTCL tumor antigen se33-1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.016278 (rank : 23) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
FSTL4_HUMAN
|
||||||
| NC score | 0.013963 (rank : 24) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6MZW2, Q8TBU0, Q9UPU1 | Gene names | FSTL4, KIAA1061 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Follistatin-related protein 4 precursor (Follistatin-like 4). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.013436 (rank : 25) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
NCK2_HUMAN
|
||||||
| NC score | 0.012285 (rank : 26) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43639, Q9BWN9, Q9UIC3 | Gene names | NCK2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoplasmic protein NCK2 (NCK adaptor protein 2) (SH2/SH3 adaptor protein NCK-beta) (Nck-2). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.011802 (rank : 27) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.009982 (rank : 28) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
DSRAD_MOUSE
|
||||||
| NC score | 0.009892 (rank : 29) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99MU3, O70375, Q80UZ6, Q8C222, Q99MU2, Q99MU4, Q99MU7 | Gene names | Adar | |||
|
Domain Architecture |

|
|||||
| Description | Double-stranded RNA-specific adenosine deaminase (EC 3.5.4.-) (DRADA) (RNA adenosine deaminase 1). | |||||
|
CAC1E_HUMAN
|
||||||
| NC score | 0.009586 (rank : 30) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15878, Q14580, Q14581 | Gene names | CACNA1E, CACH6, CACNL1A6 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent R-type calcium channel subunit alpha-1E (Voltage- gated calcium channel subunit alpha Cav2.3) (Calcium channel, L type, alpha-1 polypeptide, isoform 6) (Brain calcium channel II) (BII). | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.009043 (rank : 31) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.009004 (rank : 32) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
LYST_HUMAN
|
||||||
| NC score | 0.008602 (rank : 33) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99698, O43274, Q5T2U9, Q96TD7, Q96TD8, Q99709, Q9H133 | Gene names | LYST, CHS, CHS1 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal-trafficking regulator (Beige homolog). | |||||
|
LRTM2_HUMAN
|
||||||
| NC score | 0.006586 (rank : 34) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43300, Q7L770 | Gene names | LRRTM2, KIAA0416, LRRN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat transmembrane neuronal protein 2 precursor (Leucine-rich repeat neuronal 2 protein). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.006525 (rank : 35) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
LATS1_MOUSE
|
||||||
| NC score | 0.005030 (rank : 36) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1027 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BYR2, Q9Z0W4 | Gene names | Lats1, Warts | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase LATS1 (EC 2.7.11.1) (Large tumor suppressor homolog 1) (WARTS protein kinase). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.002377 (rank : 37) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||