Please be patient as the page loads
|
CCNT1_HUMAN
|
||||||
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |
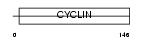
|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCNT1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.992383 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| θ value | 1.02182e-136 (rank : 3) | NC score | 0.957187 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNK_HUMAN
|
||||||
| θ value | 2.96089e-35 (rank : 4) | NC score | 0.814114 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 1.12514e-34 (rank : 5) | NC score | 0.703228 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| θ value | 8.36355e-22 (rank : 6) | NC score | 0.698498 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL2_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 7) | NC score | 0.689917 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNL1_HUMAN
|
||||||
| θ value | 4.58923e-20 (rank : 8) | NC score | 0.689911 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNL1_MOUSE
|
||||||
| θ value | 4.58923e-20 (rank : 9) | NC score | 0.690705 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNC_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 10) | NC score | 0.617258 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |
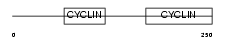
|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 11) | NC score | 0.596270 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |
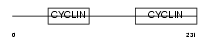
|
|||||
| Description | Cyclin-C. | |||||
|
DJBP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.066484 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
CCNA2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.096760 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 14) | NC score | 0.041181 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CD2L6_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 15) | NC score | 0.015123 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWU1, Q5JQZ7, Q5JR00, Q8TC78, Q9UPX2 | Gene names | CDC2L6, CDK11, KIAA1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6) (Death-preventing kinase) (Cyclin-dependent kinase 11). | |||||
|
CCNH_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.281074 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |
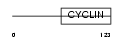
|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CD2L6_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.012882 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWD8, Q80TM1 | Gene names | Cdc2l6, Kiaa1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6). | |||||
|
ZN318_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.032712 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.092112 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.062616 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNH_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.248695 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-H. | |||||
|
UD110_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.015997 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAW8, O00474, Q6NT91 | Gene names | UGT1A10, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-10 precursor (EC 2.4.1.17) (UDPGT) (UGT1*10) (UGT1-10) (UGT1.10) (UGT-1J) (UGT1J). | |||||
|
ATF7_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.035125 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.019506 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
GBP4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.011688 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
NEO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.007230 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.009858 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
PAPPA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.030738 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
SH3K1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.010324 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
NEO1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.006348 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.038262 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PTPRV_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.007372 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
RHG06_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.008575 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.013510 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
PAPPA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.024664 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.003825 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
SEMG1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.023768 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
SRBP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.009640 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
ASCL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.008191 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
CREB5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.015584 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.002922 (rank : 49) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.002986 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
LGP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.006653 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.011349 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.012091 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SDPR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.008548 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
CCNA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.065947 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.061239 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.057263 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCNT1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O60563, O60581 | Gene names | CCNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT1_MOUSE
|
||||||
| NC score | 0.992383 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9QWV9, Q9Z0U7 | Gene names | Ccnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T1 (CycT1) (Cyclin-T). | |||||
|
CCNT2_HUMAN
|
||||||
| NC score | 0.957187 (rank : 3) | θ value | 1.02182e-136 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O60583, O60582, Q5I1Y0 | Gene names | CCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-T2 (CycT2). | |||||
|
CCNK_HUMAN
|
||||||
| NC score | 0.814114 (rank : 4) | θ value | 2.96089e-35 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75909, Q96B63, Q9NNY9 | Gene names | CCNK, CPR4 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-K. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.703228 (rank : 5) | θ value | 1.12514e-34 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CCNL2_MOUSE
|
||||||
| NC score | 0.698498 (rank : 6) | θ value | 8.36355e-22 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JJA7, Q5XK66, Q60995, Q8C136, Q8CIJ8, Q99L73, Q9QXH5 | Gene names | Ccnl2, Ania6b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Cyclin Ania-6b) (Paneth cell-enhanced expression protein) (PCEE). | |||||
|
CCNL1_MOUSE
|
||||||
| NC score | 0.690705 (rank : 7) | θ value | 4.58923e-20 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q52KE7, Q8BQ75, Q8R5H9, Q922K0, Q9CSZ3, Q9WV44 | Gene names | Ccnl1, Ania6a, Ccn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L) (Cyclin Ania-6a). | |||||
|
CCNL2_HUMAN
|
||||||
| NC score | 0.689917 (rank : 8) | θ value | 1.5773e-20 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96S94, Q5T2N5, Q5T2N6, Q6IQ12, Q7Z4Z8, Q8N3C9, Q8N3D5, Q8NHE3, Q8TEL0, Q96B00 | Gene names | CCNL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L2 (Paneth cell-enhanced expression protein). | |||||
|
CCNL1_HUMAN
|
||||||
| NC score | 0.689911 (rank : 9) | θ value | 4.58923e-20 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UK58, Q6NVY9, Q6UWS7, Q8NI48, Q96QT0, Q9NZF3 | Gene names | CCNL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-L1 (Cyclin-L). | |||||
|
CCNC_HUMAN
|
||||||
| NC score | 0.617258 (rank : 10) | θ value | 1.74796e-11 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P24863, Q9H543 | Gene names | CCNC | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNC_MOUSE
|
||||||
| NC score | 0.596270 (rank : 11) | θ value | 9.59137e-10 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62447, Q9WUZ4 | Gene names | Ccnc | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-C. | |||||
|
CCNH_HUMAN
|
||||||
| NC score | 0.281074 (rank : 12) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51946, Q8TBL9 | Gene names | CCNH | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H (MO15-associated protein) (p37) (p34). | |||||
|
CCNH_MOUSE
|
||||||
| NC score | 0.248695 (rank : 13) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61458, Q9CVJ0, Q9JHV7 | Gene names | Ccnh | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-H. | |||||
|
CCNA2_HUMAN
|
||||||
| NC score | 0.096760 (rank : 14) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20248 | Gene names | CCNA2, CCN1, CCNA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.092112 (rank : 15) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
DJBP_HUMAN
|
||||||
| NC score | 0.066484 (rank : 16) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5THR3, Q5U5T6, Q9BY88, Q9H5C4, Q9NSF5 | Gene names | DJBP, KIAA1672 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DJ-1-binding protein (CAP-binding protein complex-interacting protein 1). | |||||
|
CCNA1_HUMAN
|
||||||
| NC score | 0.065947 (rank : 17) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78396 | Gene names | CCNA1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.062616 (rank : 18) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CCNA1_MOUSE
|
||||||
| NC score | 0.061239 (rank : 19) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q61456 | Gene names | Ccna1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A1. | |||||
|
CCNA2_MOUSE
|
||||||
| NC score | 0.057263 (rank : 20) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P51943, Q61459 | Gene names | Ccna2, Ccna, Cyca | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-A2 (Cyclin-A). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.041181 (rank : 21) | θ value | 0.0330416 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.038262 (rank : 22) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ATF7_HUMAN
|
||||||
| NC score | 0.035125 (rank : 23) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17544, Q13814 | Gene names | ATF7, ATFA | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-7 (Activating transcription factor 7) (Transcription factor ATF-A). | |||||
|
ZN318_MOUSE
|
||||||
| NC score | 0.032712 (rank : 24) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 642 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PP2, Q3TZL5, Q9JJ01 | Gene names | Znf318, Tzf, Zfp318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Testicular zinc finger protein). | |||||
|
PAPPA_MOUSE
|
||||||
| NC score | 0.030738 (rank : 25) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4K8, Q80VW3, Q80YY1, Q8K423, Q8R4K7, Q9ES06, Q9JK57 | Gene names | Pappa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
PAPPA_HUMAN
|
||||||
| NC score | 0.024664 (rank : 26) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13219, Q08371, Q68G52, Q9UDK7 | Gene names | PAPPA | |||
|
Domain Architecture |

|
|||||
| Description | Pappalysin-1 precursor (EC 3.4.24.79) (Pregnancy-associated plasma protein-A) (PAPP-A) (Insulin-like growth factor-dependent IGF-binding protein 4 protease) (IGF-dependent IGFBP-4 protease) (IGFBP-4ase). | |||||
|
SEMG1_HUMAN
|
||||||
| NC score | 0.023768 (rank : 27) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04279, Q6X4I9, Q6Y809, Q6Y822, Q6Y823, Q86U64, Q96QM3 | Gene names | SEMG1, SEMG | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-1 precursor (Semenogelin I) (SGI) [Contains: Alpha- inhibin-92; Alpha-inhibin-31; Seminal basic protein]. | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.019506 (rank : 28) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
UD110_HUMAN
|
||||||
| NC score | 0.015997 (rank : 29) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HAW8, O00474, Q6NT91 | Gene names | UGT1A10, GNT1, UGT1 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-glucuronosyltransferase 1-10 precursor (EC 2.4.1.17) (UDPGT) (UGT1*10) (UGT1-10) (UGT1.10) (UGT-1J) (UGT1J). | |||||
|
CREB5_HUMAN
|
||||||
| NC score | 0.015584 (rank : 30) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q02930, Q05886, Q06246 | Gene names | CREB5, CREBPA | |||
|
Domain Architecture |

|
|||||
| Description | cAMP response element-binding protein 5 (CRE-BPa). | |||||
|
CD2L6_HUMAN
|
||||||
| NC score | 0.015123 (rank : 31) | θ value | 0.0563607 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BWU1, Q5JQZ7, Q5JR00, Q8TC78, Q9UPX2 | Gene names | CDC2L6, CDK11, KIAA1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6) (Death-preventing kinase) (Cyclin-dependent kinase 11). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.013510 (rank : 32) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CD2L6_MOUSE
|
||||||
| NC score | 0.012882 (rank : 33) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BWD8, Q80TM1 | Gene names | Cdc2l6, Kiaa1028 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell division cycle 2-like protein kinase 6 (EC 2.7.11.22) (CDC2- related protein kinase 6). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.012091 (rank : 34) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
GBP4_HUMAN
|
||||||
| NC score | 0.011688 (rank : 35) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96PP9, Q86T99 | Gene names | GBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylate-binding protein 4 (GTP-binding protein 4) (Guanine nucleotide-binding protein 4) (GBP-4). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.011349 (rank : 36) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SH3K1_MOUSE
|
||||||
| NC score | 0.010324 (rank : 37) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R550, Q8CAL8, Q8CEF6, Q8R545, Q8R546, Q8R547, Q8R548, Q8R549, Q8R551, Q9CTQ9, Q9DC14, Q9JKC3 | Gene names | Sh3kbp1, Ruk, Seta | |||
|
Domain Architecture |

|
|||||
| Description | SH3-domain kinase-binding protein 1 (SH3-containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk). | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.009858 (rank : 38) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
SRBP1_MOUSE
|
||||||
| NC score | 0.009640 (rank : 39) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9WTN3 | Gene names | Srebf1, Srebp1 | |||
|
Domain Architecture |

|
|||||
| Description | Sterol regulatory element-binding protein 1 (SREBP-1) (Sterol regulatory element-binding transcription factor 1). | |||||
|
RHG06_MOUSE
|
||||||
| NC score | 0.008575 (rank : 40) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O54834, Q9QZL8 | Gene names | Arhgap6 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-GTPase-activating protein 6 (Rho-type GTPase-activating protein RhoGAPX-1). | |||||
|
SDPR_HUMAN
|
||||||
| NC score | 0.008548 (rank : 41) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95810 | Gene names | SDPR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein) (PS-p68). | |||||
|
ASCL1_MOUSE
|
||||||
| NC score | 0.008191 (rank : 42) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q02067 | Gene names | Ascl1, Ash1, Mash-1, Mash1 | |||
|
Domain Architecture |

|
|||||
| Description | Achaete-scute homolog 1 (Mash-1). | |||||
|
PTPRV_MOUSE
|
||||||
| NC score | 0.007372 (rank : 43) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70289 | Gene names | Ptprv, Esp | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase V precursor (EC 3.1.3.48) (Embryonic stem cell protein-tyrosine phosphatase) (ES cell phosphatase). | |||||
|
NEO1_HUMAN
|
||||||
| NC score | 0.007230 (rank : 44) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92859, O00340 | Gene names | NEO1, NGN | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
LGP2_MOUSE
|
||||||
| NC score | 0.006653 (rank : 45) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99J87, Q9D1X4 | Gene names | Lgp2, D11lgp2e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent helicase LGP2 (EC 3.6.1.-) (Protein D11Lgp2). | |||||
|
NEO1_MOUSE
|
||||||
| NC score | 0.006348 (rank : 46) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97798 | Gene names | Neo1, Ngn | |||
|
Domain Architecture |

|
|||||
| Description | Neogenin precursor. | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.003825 (rank : 47) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.002986 (rank : 48) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.002922 (rank : 49) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||