Please be patient as the page loads
|
TMUB1_MOUSE
|
||||||
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |
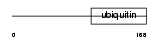
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TMUB1_MOUSE
|
||||||
| θ value | 2.02215e-84 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
|
TMUB1_HUMAN
|
||||||
| θ value | 1.60597e-73 (rank : 2) | NC score | 0.974828 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |
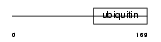
|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
TMUB2_MOUSE
|
||||||
| θ value | 5.22648e-32 (rank : 3) | NC score | 0.852448 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
TMUB2_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 4) | NC score | 0.849896 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71RG4, Q8NDI2, Q9BPZ5, Q9HAG3 | Gene names | TMUB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.049363 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.048819 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CABL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.039378 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 8) | NC score | 0.023824 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.010776 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
PPHLN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.038523 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEY8, Q86YT2, Q8IXN3, Q8TB09, Q96NB9, Q9NXL4, Q9P0P6, Q9P0R9 | Gene names | PPHLN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1 (Gastric cancer antigen Ga50). | |||||
|
CER1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.031851 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95813, Q6ISJ1, Q6ISJ6, Q6ISQ2, Q6ISS1 | Gene names | CER1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerberus precursor (Cerberus-related protein). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.035279 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
HDAC7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.015028 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
NIBL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.027104 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.026654 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
ZMYM3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.020757 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
ZN687_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.007913 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
GRASP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.017312 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
LRC50_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.020397 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.016914 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.006052 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IGHG4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 22) | NC score | 0.010089 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01861 | Gene names | IGHG4 | |||
|
Domain Architecture |
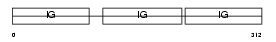
|
|||||
| Description | Ig gamma-4 chain C region. | |||||
|
NEB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.016971 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.020914 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.012638 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
RFIP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.016192 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.028541 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
INAR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.018298 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35664, O35238, O35663, O35983 | Gene names | Ifnar2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.016095 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
NCOR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.016357 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.005237 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
SON_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.042507 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.016963 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.023438 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
TMUB1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.02215e-84 (rank : 1) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JMG3 | Gene names | Tmub1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1. | |||||
|
TMUB1_HUMAN
|
||||||
| NC score | 0.974828 (rank : 2) | θ value | 1.60597e-73 (rank : 2) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9BVT8 | Gene names | TMUB1, C7orf21 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 1 (Ubiquitin-like protein SB144). | |||||
|
TMUB2_MOUSE
|
||||||
| NC score | 0.852448 (rank : 3) | θ value | 5.22648e-32 (rank : 3) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3V209, Q8K0X0, Q91YQ0, Q9D8C5 | Gene names | Tmub2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
TMUB2_HUMAN
|
||||||
| NC score | 0.849896 (rank : 4) | θ value | 1.73987e-27 (rank : 4) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q71RG4, Q8NDI2, Q9BPZ5, Q9HAG3 | Gene names | TMUB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and ubiquitin-like domain-containing protein 2. | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.049363 (rank : 5) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.048819 (rank : 6) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.042507 (rank : 7) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CABL1_HUMAN
|
||||||
| NC score | 0.039378 (rank : 8) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TDN4, Q8N3Y8, Q8NA22, Q9BTG1 | Gene names | CABLES1, CABLES | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 and ABL1 enzyme substrate 1 (Interactor with CDK3 1) (Ik3-1). | |||||
|
PPHLN_HUMAN
|
||||||
| NC score | 0.038523 (rank : 9) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NEY8, Q86YT2, Q8IXN3, Q8TB09, Q96NB9, Q9NXL4, Q9P0P6, Q9P0R9 | Gene names | PPHLN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Periphilin-1 (Gastric cancer antigen Ga50). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.035279 (rank : 10) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
CER1_HUMAN
|
||||||
| NC score | 0.031851 (rank : 11) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95813, Q6ISJ1, Q6ISJ6, Q6ISQ2, Q6ISS1 | Gene names | CER1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerberus precursor (Cerberus-related protein). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.028541 (rank : 12) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
NIBL_HUMAN
|
||||||
| NC score | 0.027104 (rank : 13) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96TA1, Q9BUS2, Q9NT35 | Gene names | C9orf88 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban-like protein (Meg-3). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.026654 (rank : 14) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.023824 (rank : 15) | θ value | 1.06291 (rank : 8) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.023438 (rank : 16) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.020914 (rank : 17) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
ZMYM3_HUMAN
|
||||||
| NC score | 0.020757 (rank : 18) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14202, O15089 | Gene names | ZMYM3, DXS6673E, KIAA0385, ZNF261 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYM-type protein 3 (Zinc finger protein 261). | |||||
|
LRC50_HUMAN
|
||||||
| NC score | 0.020397 (rank : 19) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NEP3, Q69YI8, Q69YJ0, Q96LP3 | Gene names | LRRC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 50. | |||||
|
INAR2_MOUSE
|
||||||
| NC score | 0.018298 (rank : 20) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35664, O35238, O35663, O35983 | Gene names | Ifnar2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interferon-alpha/beta receptor beta chain precursor (IFN-alpha-REC) (Type I interferon receptor) (IFN-R) (Interferon alpha/beta receptor 2). | |||||
|
GRASP_MOUSE
|
||||||
| NC score | 0.017312 (rank : 21) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JJA9, Q9JKL0 | Gene names | Grasp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | General receptor for phosphoinositides 1-associated scaffold protein (GRP1-associated scaffold protein). | |||||
|
NEB2_MOUSE
|
||||||
| NC score | 0.016971 (rank : 22) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6R891, Q8K0X7 | Gene names | Ppp1r9b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.016963 (rank : 23) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.016914 (rank : 24) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
NCOR1_MOUSE
|
||||||
| NC score | 0.016357 (rank : 25) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60974, Q60812 | Gene names | Ncor1, Rxrip13 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR) (Retinoid X receptor- interacting protein 13) (RIP13). | |||||
|
RFIP5_MOUSE
|
||||||
| NC score | 0.016192 (rank : 26) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R361 | Gene names | Rab11fip5, D6Ertd32e, Rip11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.016095 (rank : 27) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
HDAC7_MOUSE
|
||||||
| NC score | 0.015028 (rank : 28) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C2B3, Q8C2C9, Q8C8X4, Q8CB80, Q8CDA3, Q9JL72 | Gene names | Hdac7a, Hdac7 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 7a (HD7a). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.012638 (rank : 29) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.010776 (rank : 30) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
IGHG4_HUMAN
|
||||||
| NC score | 0.010089 (rank : 31) | θ value | 6.88961 (rank : 22) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01861 | Gene names | IGHG4 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-4 chain C region. | |||||
|
ZN687_MOUSE
|
||||||
| NC score | 0.007913 (rank : 32) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 913 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D2D7, Q6PAP3, Q6ZPQ9 | Gene names | Znf687, Kiaa1441, Zfp687 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 687. | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.006052 (rank : 33) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.005237 (rank : 34) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 34 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||