Please be patient as the page loads
|
JHD3A_HUMAN
|
||||||
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
JHD3A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998809 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.987431 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 1.31915e-176 (rank : 4) | NC score | 0.980163 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 6.12769e-174 (rank : 5) | NC score | 0.972314 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 1.36511e-173 (rank : 6) | NC score | 0.968156 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | 8.33966e-147 (rank : 7) | NC score | 0.884716 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_MOUSE
|
||||||
| θ value | 2.05413e-145 (rank : 8) | NC score | 0.886936 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 9) | NC score | 0.624933 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 10) | NC score | 0.615767 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 11) | NC score | 0.616975 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | 4.00176e-32 (rank : 12) | NC score | 0.615616 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 13) | NC score | 0.627534 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 1.33217e-27 (rank : 14) | NC score | 0.585622 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 2.27234e-27 (rank : 15) | NC score | 0.599101 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 1.99067e-23 (rank : 16) | NC score | 0.563123 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
AF17_HUMAN
|
||||||
| θ value | 2.43343e-21 (rank : 17) | NC score | 0.661925 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 4.15078e-21 (rank : 18) | NC score | 0.587161 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 19) | NC score | 0.557310 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
AF10_HUMAN
|
||||||
| θ value | 7.0802e-21 (rank : 20) | NC score | 0.629651 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_MOUSE
|
||||||
| θ value | 7.0802e-21 (rank : 21) | NC score | 0.630878 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
JADE1_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 22) | NC score | 0.592873 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 23) | NC score | 0.591961 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 24) | NC score | 0.596694 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 1.13037e-18 (rank : 25) | NC score | 0.595792 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 26) | NC score | 0.581636 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 3.28887e-18 (rank : 27) | NC score | 0.593323 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 28) | NC score | 0.542632 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 29) | NC score | 0.540888 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 30) | NC score | 0.184792 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 31) | NC score | 0.170537 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 32) | NC score | 0.165442 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 33) | NC score | 0.175731 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 34) | NC score | 0.162390 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
LBR_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 35) | NC score | 0.111571 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14739, Q14740, Q53GU7, Q59FE6 | Gene names | LBR | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein) (LMN2R). | |||||
|
LBR_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 36) | NC score | 0.108421 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3U9G9, Q3TSW2, Q811V8, Q811V9, Q8BST3, Q8K2Y8, Q8VDM0, Q91YS5, Q91Z27 | Gene names | Lbr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 37) | NC score | 0.187020 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 38) | NC score | 0.116460 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
PHF11_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.192785 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF7_HUMAN
|
||||||
| θ value | 0.47712 (rank : 40) | NC score | 0.143698 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
CRDL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.019648 (rank : 116) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEA6, Q925I3 | Gene names | Chrdl2, Chl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor. | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.152866 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.154123 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
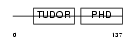
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.055069 (rank : 108) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
RECK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.027174 (rank : 114) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
SYP2L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.015037 (rank : 121) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.018414 (rank : 117) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.014793 (rank : 122) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
CLCN6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.007934 (rank : 131) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35454 | Gene names | Clcn6, Clc6 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 6 (ClC-6). | |||||
|
PF21B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.097761 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RECK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.023547 (rank : 115) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.084324 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.083470 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
DHRS9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.011892 (rank : 126) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58NB6, Q8BGC7 | Gene names | Dhrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (Short-chain dehydrogenase/reductase retSDR8). | |||||
|
ITA4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.011473 (rank : 128) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00651 | Gene names | Itga4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (Lymphocyte Peyer patch adhesion molecules subunit alpha) (LPAM subunit alpha) (CD49d antigen). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.099047 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
RBM10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.008995 (rank : 129) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
SPY2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011671 (rank : 127) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
SPY2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.012341 (rank : 124) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.090768 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.027333 (rank : 113) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.029591 (rank : 112) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.018292 (rank : 118) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CR025_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.011942 (rank : 125) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
DSCL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.000164 (rank : 137) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.121667 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SRBS1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.007526 (rank : 133) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
TAF1C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.017461 (rank : 120) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
TEP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.007315 (rank : 134) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.002476 (rank : 135) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
ATPG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.014012 (rank : 123) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VR2 | Gene names | Atp5c1 | |||
|
Domain Architecture |
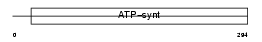
|
|||||
| Description | ATP synthase gamma chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.083293 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CJ084_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.008743 (rank : 130) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W3, Q9H5V5 | Gene names | C10orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf84. | |||||
|
K1333_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.088723 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
NIP30_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.007699 (rank : 132) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZU8 | Gene names | NIP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
NU153_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.017827 (rank : 119) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.000973 (rank : 136) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
ZNF18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | -0.004253 (rank : 138) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17022, Q5QHQ3, Q8IYC4, Q8NAH6 | Gene names | ZNF18, HDSG1, KOX11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein KOX11) (Heart development- specific gene 1 protein). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.154080 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.146119 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.055608 (rank : 106) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.060833 (rank : 103) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ARI3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.102291 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
ARI3A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.105332 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
ARI4A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.081657 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
ARI5B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.062073 (rank : 102) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
ARI5B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.065065 (rank : 98) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
ARID2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.078459 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.122588 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.120601 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.131777 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.105863 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.105358 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.100727 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.095602 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
BRD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.054256 (rank : 109) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.053029 (rank : 110) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.055444 (rank : 107) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.052986 (rank : 111) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
CHD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.088815 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.111125 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.118873 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
K1333_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.064464 (rank : 99) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
MTF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.133957 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
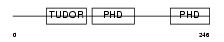
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.137436 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.094953 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.091477 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.092933 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.096140 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.095802 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.104398 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.118249 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.117450 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
PHF10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.159027 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.159278 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.097115 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.095815 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.083698 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.076530 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.084085 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
REQU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.113338 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.111622 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.124582 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.089771 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.071526 (rank : 93) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.072009 (rank : 92) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.066928 (rank : 96) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.064048 (rank : 100) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.063066 (rank : 101) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.066692 (rank : 97) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.058118 (rank : 105) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.058708 (rank : 104) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TRI66_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.067719 (rank : 95) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.069051 (rank : 94) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.085674 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.080092 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.092372 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.098123 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.998809 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.987431 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.980163 (rank : 4) | θ value | 1.31915e-176 (rank : 4) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.972314 (rank : 5) | θ value | 6.12769e-174 (rank : 5) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.968156 (rank : 6) | θ value | 1.36511e-173 (rank : 6) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3D_MOUSE
|
||||||
| NC score | 0.886936 (rank : 7) | θ value | 2.05413e-145 (rank : 8) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.884716 (rank : 8) | θ value | 8.33966e-147 (rank : 7) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.661925 (rank : 9) | θ value | 2.43343e-21 (rank : 17) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.630878 (rank : 10) | θ value | 7.0802e-21 (rank : 21) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.629651 (rank : 11) | θ value | 7.0802e-21 (rank : 20) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.627534 (rank : 12) | θ value | 5.22648e-32 (rank : 13) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.624933 (rank : 13) | θ value | 2.50655e-34 (rank : 9) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.616975 (rank : 14) | θ value | 3.06405e-32 (rank : 11) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.615767 (rank : 15) | θ value | 3.06405e-32 (rank : 10) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.615616 (rank : 16) | θ value | 4.00176e-32 (rank : 12) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.599101 (rank : 17) | θ value | 2.27234e-27 (rank : 15) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.596694 (rank : 18) | θ value | 6.62687e-19 (rank : 24) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.595792 (rank : 19) | θ value | 1.13037e-18 (rank : 25) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.593323 (rank : 20) | θ value | 3.28887e-18 (rank : 27) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.592873 (rank : 21) | θ value | 2.27762e-19 (rank : 22) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.591961 (rank : 22) | θ value | 2.97466e-19 (rank : 23) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.587161 (rank : 23) | θ value | 4.15078e-21 (rank : 18) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.585622 (rank : 24) | θ value | 1.33217e-27 (rank : 14) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.581636 (rank : 25) | θ value | 3.28887e-18 (rank : 26) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.563123 (rank : 26) | θ value | 1.99067e-23 (rank : 16) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.557310 (rank : 27) | θ value | 4.15078e-21 (rank : 19) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.542632 (rank : 28) | θ value | 1.68911e-14 (rank : 28) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.540888 (rank : 29) | θ value | 3.18553e-13 (rank : 29) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.192785 (rank : 30) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.187020 (rank : 31) | θ value | 0.0563607 (rank : 37) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.184792 (rank : 32) | θ value | 0.00134147 (rank : 30) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.175731 (rank : 33) | θ value | 0.00509761 (rank : 33) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.170537 (rank : 34) | θ value | 0.00228821 (rank : 31) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.165442 (rank : 35) | θ value | 0.00390308 (rank : 32) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.162390 (rank : 36) | θ value | 0.0113563 (rank : 34) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.159278 (rank : 37) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.159027 (rank : 38) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.154123 (rank : 39) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.154080 (rank : 40) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.152866 (rank : 41) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.146119 (rank : 42) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.143698 (rank : 43) | θ value | 0.47712 (rank : 40) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MTF2_MOUSE
|
||||||
| NC score | 0.137436 (rank : 44) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.133957 (rank : 45) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.131777 (rank : 46) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.124582 (rank : 47) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.122588 (rank : 48) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.121667 (rank : 49) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.120601 (rank : 50) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.118873 (rank : 51) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.118249 (rank : 52) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.117450 (rank : 53) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.116460 (rank : 54) | θ value | 0.125558 (rank : 38) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.113338 (rank : 55) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.111622 (rank : 56) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
LBR_HUMAN
|
||||||
| NC score | 0.111571 (rank : 57) | θ value | 0.0113563 (rank : 35) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14739, Q14740, Q53GU7, Q59FE6 | Gene names | LBR | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein) (LMN2R). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.111125 (rank : 58) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
LBR_MOUSE
|
||||||
| NC score | 0.108421 (rank : 59) | θ value | 0.0193708 (rank : 36) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q3U9G9, Q3TSW2, Q811V8, Q811V9, Q8BST3, Q8K2Y8, Q8VDM0, Q91YS5, Q91Z27 | Gene names | Lbr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lamin-B receptor (Integral nuclear envelope inner membrane protein). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.105863 (rank : 60) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.105358 (rank : 61) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
ARI3A_MOUSE
|
||||||
| NC score | 0.105332 (rank : 62) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q62431 | Gene names | Arid3a, Dri1, Dril1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (Dead ringer-like protein 1) (B-cell regulator of IgH transcription) (Bright). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.104398 (rank : 63) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
ARI3A_HUMAN
|
||||||
| NC score | 0.102291 (rank : 64) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99856, Q8IZA7 | Gene names | ARID3A, DRIL1, DRX, E2FBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 3A (ARID domain- containing protein 3A) (B-cell regulator of IgH transcription) (Bright) (E2F-binding protein 1). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.100727 (rank : 65) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.099047 (rank : 66) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.098123 (rank : 67) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.097761 (rank : 68) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.097115 (rank : 69) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.096140 (rank : 70) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.095815 (rank : 71) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.095802 (rank : 72) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.095602 (rank : 73) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.094953 (rank : 74) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.092933 (rank : 75) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.092372 (rank : 76) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.091477 (rank : 77) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.090768 (rank : 78) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.089771 (rank : 79) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.088815 (rank : 80) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.088723 (rank : 81) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.085674 (rank : 82) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.084324 (rank : 83) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.084085 (rank : 84) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.083698 (rank : 85) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.083470 (rank : 86) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.083293 (rank : 87) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
ARI4A_HUMAN
|
||||||
| NC score | 0.081657 (rank : 88) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P29374, Q15991, Q15992, Q15993 | Gene names | ARID4A, RBBP1, RBP1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 4A (ARID domain- containing protein 4A) (Retinoblastoma-binding protein 1) (RBBP-1). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.080092 (rank : 89) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
ARID2_HUMAN
|
||||||
| NC score | 0.078459 (rank : 90) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q68CP9, Q5EB51, Q645I3, Q6ZRY5, Q7Z3I5, Q86T28, Q96SJ6, Q9HCL5 | Gene names | ARID2, KIAA1557 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 2 (ARID domain- containing protein 2) (BRG1-associated factor 200) (BAF200). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.076530 (rank : 91) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.072009 (rank : 92) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.071526 (rank : 93) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.069051 (rank : 94) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.067719 (rank : 95) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.066928 (rank : 96) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.066692 (rank : 97) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
ARI5B_MOUSE
|
||||||
| NC score | 0.065065 (rank : 98) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BM75, Q8C0E0, Q8K4G8, Q8K4L9, Q9CU78, Q9JIX4 | Gene names | Arid5b, Desrt, Mrf2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (MRF1-like) (Modulator recognition factor protein 2) (MRF-2) (Developmentally and sexually retarded with transient immune abnormalities protein). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.064464 (rank : 99) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.064048 (rank : 100) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.063066 (rank : 101) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
ARI5B_HUMAN
|
||||||
| NC score | 0.062073 (rank : 102) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14865, Q7Z3M4, Q8N421, Q9H786 | Gene names | ARID5B, DESRT, MRF2 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 5B (ARID domain- containing protein 5B) (Mrf1-like) (Modulator recognition factor 2) (MRF-2). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.060833 (rank : 103) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.058708 (rank : 104) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.058118 (rank : 105) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.055608 (rank : 106) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
BRD9_HUMAN
|
||||||
| NC score | 0.055444 (rank : 107) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.055069 (rank : 108) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.054256 (rank : 109) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.053029 (rank : 110) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.052986 (rank : 111) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.029591 (rank : 112) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.027333 (rank : 113) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
RECK_MOUSE
|
||||||
| NC score | 0.027174 (rank : 114) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z0J1 | Gene names | Reck | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (mRECK). | |||||
|
RECK_HUMAN
|
||||||
| NC score | 0.023547 (rank : 115) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95980, Q8WX37 | Gene names | RECK | |||
|
Domain Architecture |

|
|||||
| Description | Reversion-inducing cysteine-rich protein with Kazal motifs precursor (hRECK) (Suppressor of tumorigenicity 15) (ST15). | |||||
|
CRDL2_MOUSE
|
||||||
| NC score | 0.019648 (rank : 116) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VEA6, Q925I3 | Gene names | Chrdl2, Chl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chordin-like protein 2 precursor. | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.018414 (rank : 117) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.018292 (rank : 118) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NU153_HUMAN
|
||||||
| NC score | 0.017827 (rank : 119) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P49790 | Gene names | NUP153 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup153 (Nucleoporin Nup153) (153 kDa nucleoporin). | |||||
|
TAF1C_MOUSE
|
||||||
| NC score | 0.017461 (rank : 120) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PDZ2, P97359, Q3U3B6, Q8BN54 | Gene names | Taf1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA box-binding protein-associated factor, RNA polymerase I, subunit C (TATA box-binding protein-associated factor 1C) (TBP-associated factor 1C) (TBP-associated factor RNA polymerase I 95 kDa) (TAFI95). | |||||
|
SYP2L_HUMAN
|
||||||
| NC score | 0.015037 (rank : 121) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H987 | Gene names | SYNPO2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.014793 (rank : 122) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
ATPG_MOUSE
|
||||||
| NC score | 0.014012 (rank : 123) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VR2 | Gene names | Atp5c1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP synthase gamma chain, mitochondrial precursor (EC 3.6.3.14). | |||||
|
SPY2_MOUSE
|
||||||
| NC score | 0.012341 (rank : 124) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXV8, Q9WUQ9 | Gene names | Spry2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
CR025_HUMAN
|
||||||
| NC score | 0.011942 (rank : 125) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
DHRS9_MOUSE
|
||||||
| NC score | 0.011892 (rank : 126) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q58NB6, Q8BGC7 | Gene names | Dhrs9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dehydrogenase/reductase SDR family member 9 precursor (EC 1.1.-.-) (3- alpha hydroxysteroid dehydrogenase) (3alpha-HSD) (Short-chain dehydrogenase/reductase retSDR8). | |||||
|
SPY2_HUMAN
|
||||||
| NC score | 0.011671 (rank : 127) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43597, Q5T6Z7 | Gene names | SPRY2 | |||
|
Domain Architecture |

|
|||||
| Description | Sprouty homolog 2 (Spry-2). | |||||
|
ITA4_MOUSE
|
||||||
| NC score | 0.011473 (rank : 128) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00651 | Gene names | Itga4 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-4 precursor (Integrin alpha-IV) (VLA-4) (Lymphocyte Peyer patch adhesion molecules subunit alpha) (LPAM subunit alpha) (CD49d antigen). | |||||
|
RBM10_HUMAN
|
||||||
| NC score | 0.008995 (rank : 129) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98175, Q14136 | Gene names | RBM10, DXS8237E, KIAA0122 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 10 (RNA-binding motif protein 10). | |||||
|
CJ084_HUMAN
|
||||||
| NC score | 0.008743 (rank : 130) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W3, Q9H5V5 | Gene names | C10orf84 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf84. | |||||
|
CLCN6_MOUSE
|
||||||
| NC score | 0.007934 (rank : 131) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35454 | Gene names | Clcn6, Clc6 | |||
|
Domain Architecture |

|
|||||
| Description | Chloride channel protein 6 (ClC-6). | |||||
|
NIP30_HUMAN
|
||||||
| NC score | 0.007699 (rank : 132) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9GZU8 | Gene names | NIP30 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NEFA-interacting nuclear protein NIP30. | |||||
|
SRBS1_MOUSE
|
||||||
| NC score | 0.007526 (rank : 133) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62417, Q80TF8, Q8BZI3, Q8K3Y2, Q921F8, Q9Z0Z8, Q9Z0Z9 | Gene names | Sorbs1, Kiaa1296, Sh3d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
TEP1_HUMAN
|
||||||
| NC score | 0.007315 (rank : 134) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 371 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99973 | Gene names | TEP1, TLP1, TP1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase protein component 1 (Telomerase-associated protein 1) (Telomerase protein 1) (p240) (p80 telomerase homolog). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.002476 (rank : 135) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.000973 (rank : 136) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
DSCL1_HUMAN
|
||||||
| NC score | 0.000164 (rank : 137) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TD84, Q76MU9, Q8IZY3, Q8IZY4, Q8WXU7, Q9ULT7 | Gene names | DSCAML1, DSCAM2, KIAA1132 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Down syndrome cell adhesion molecule-like protein 1 precursor (Down syndrome cell adhesion molecule 2). | |||||
|
ZNF18_HUMAN
|
||||||
| NC score | -0.004253 (rank : 138) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 78 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P17022, Q5QHQ3, Q8IYC4, Q8NAH6 | Gene names | ZNF18, HDSG1, KOX11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 18 (Zinc finger protein KOX11) (Heart development- specific gene 1 protein). | |||||