Please be patient as the page loads
|
CR025_HUMAN
|
||||||
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CR025_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
CR025_MOUSE
|
||||||
| θ value | 5.48135e-98 (rank : 2) | NC score | 0.979606 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH50, Q3TQ72, Q8BU43 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25 homolog. | |||||
|
MUC13_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 3) | NC score | 0.110156 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 4) | NC score | 0.085243 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 5) | NC score | 0.078938 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.115636 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CN037_MOUSE
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.092696 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.104864 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.028760 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.176097 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.067744 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 12) | NC score | 0.066552 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.058018 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
ZCH10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.115667 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
CD22_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.018567 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.055782 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.028060 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
TTBK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.011615 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
USBP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.054218 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.009568 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
ZFY2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.003963 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
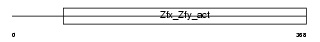
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
CF010_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.070416 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
IMA3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.034922 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |
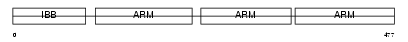
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.034856 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |
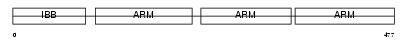
|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.029669 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.016088 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
E2F4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.024929 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.018295 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.042849 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.043457 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
AFF3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.038256 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CCD21_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.020717 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.036001 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
HAP28_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.067916 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13442, Q92906 | Gene names | PDAP1, HASPP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1). | |||||
|
HAP28_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.069421 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHX2, Q3KQJ5 | Gene names | Pdap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1). | |||||
|
ADNP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.026144 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z103 | Gene names | Adnp | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.011942 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
KIF17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.009407 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.018713 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.012621 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CHD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.012467 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
IPPD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.048044 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60829, Q91XB5 | Gene names | Ppp1r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.035267 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.011989 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.017091 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.001163 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.038307 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.014330 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
FA35A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.032137 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.049565 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.025362 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.023133 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.042009 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TR150_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.027176 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
UBP31_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.012129 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
ZN326_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.020357 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.056786 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.057832 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
CR025_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q96B23, Q5XG78, Q6N058, Q86TB5, Q8TCQ5 | Gene names | C18orf25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25. | |||||
|
CR025_MOUSE
|
||||||
| NC score | 0.979606 (rank : 2) | θ value | 5.48135e-98 (rank : 2) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BH50, Q3TQ72, Q8BU43 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf25 homolog. | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.176097 (rank : 3) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZCH10_HUMAN
|
||||||
| NC score | 0.115667 (rank : 4) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TBK6, Q9NXR4 | Gene names | ZCCHC10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.115636 (rank : 5) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MUC13_MOUSE
|
||||||
| NC score | 0.110156 (rank : 6) | θ value | 0.0193708 (rank : 3) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P19467 | Gene names | Muc13, Ly64 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-13 precursor (Cell surface antigen 114/A10) (Lymphocyte antigen 64). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.104864 (rank : 7) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
CN037_MOUSE
|
||||||
| NC score | 0.092696 (rank : 8) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BT18, Q8BFQ7, Q8R1Q5, Q9D6C5 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf37 homolog precursor. | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.085243 (rank : 9) | θ value | 0.0736092 (rank : 4) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.078938 (rank : 10) | θ value | 0.0736092 (rank : 5) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.070416 (rank : 11) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
HAP28_MOUSE
|
||||||
| NC score | 0.069421 (rank : 12) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q3UHX2, Q3KQJ5 | Gene names | Pdap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1). | |||||
|
HAP28_HUMAN
|
||||||
| NC score | 0.067916 (rank : 13) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13442, Q92906 | Gene names | PDAP1, HASPP28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 28 kDa heat- and acid-stable phosphoprotein (PDGF-associated protein) (PAP) (PDGFA-associated protein 1) (PAP1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.067744 (rank : 14) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.066552 (rank : 15) | θ value | 0.365318 (rank : 12) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.058018 (rank : 16) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.057832 (rank : 17) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.056786 (rank : 18) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.055782 (rank : 19) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
USBP1_HUMAN
|
||||||
| NC score | 0.054218 (rank : 20) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N6Y0, Q8NBX7, Q96KH3, Q9BYI8 | Gene names | USHBP1, AIEBP, MCC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USH1C-binding protein 1 (Usher syndrome type-1C protein-binding protein 1) (MCC-2) (AIE-75 binding protein). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.049565 (rank : 21) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
IPPD_MOUSE
|
||||||
| NC score | 0.048044 (rank : 22) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q60829, Q91XB5 | Gene names | Ppp1r1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dopamine- and cAMP-regulated neuronal phosphoprotein (DARPP-32). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.043457 (rank : 23) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.042849 (rank : 24) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.042009 (rank : 25) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.038307 (rank : 26) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AFF3_HUMAN
|
||||||
| NC score | 0.038256 (rank : 27) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P51826 | Gene names | AFF3, LAF4 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 3 (Protein LAF-4) (Lymphoid nuclear protein related to AF4). | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.036001 (rank : 28) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.035267 (rank : 29) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
IMA3_HUMAN
|
||||||
| NC score | 0.034922 (rank : 30) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00505, O00191, O43195, Q96AA7 | Gene names | KPNA3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (SRP1-gamma). | |||||
|
IMA3_MOUSE
|
||||||
| NC score | 0.034856 (rank : 31) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35344 | Gene names | Kpna3 | |||
|
Domain Architecture |

|
|||||
| Description | Importin alpha-3 subunit (Karyopherin alpha-3 subunit) (Importin alpha Q2). | |||||
|
FA35A_MOUSE
|
||||||
| NC score | 0.032137 (rank : 32) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UEN2 | Gene names | Fam35a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM35A. | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.029669 (rank : 33) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.028760 (rank : 34) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.028060 (rank : 35) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.027176 (rank : 36) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
ADNP_MOUSE
|
||||||
| NC score | 0.026144 (rank : 37) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z103 | Gene names | Adnp | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.025362 (rank : 38) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
E2F4_HUMAN
|
||||||
| NC score | 0.024929 (rank : 39) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16254, Q12991, Q15328 | Gene names | E2F4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F4 (E2F-4). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.023133 (rank : 40) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
CCD21_MOUSE
|
||||||
| NC score | 0.020717 (rank : 41) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BMK0, Q8BUF1, Q8K0E6 | Gene names | Ccdc21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
ZN326_MOUSE
|
||||||
| NC score | 0.020357 (rank : 42) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O88291, Q8BSJ5, Q8K1X9, Q9CYG9 | Gene names | Znf326, Zan75, Zfp326 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 326 (Zinc finger protein-associated with nuclear matrix of 75 kDa). | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.018713 (rank : 43) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
CD22_HUMAN
|
||||||
| NC score | 0.018567 (rank : 44) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20273, O95699, O95701, O95702, O95703, Q01665, Q92872, Q92873, Q9UQA6, Q9UQA7, Q9UQA8, Q9UQA9, Q9UQB0, Q9Y2A6 | Gene names | CD22 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor CD22 precursor (Leu-14) (B-lymphocyte cell adhesion molecule) (BL-CAM) (Siglec-2). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.018295 (rank : 45) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.017091 (rank : 46) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.016088 (rank : 47) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.014330 (rank : 48) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.012621 (rank : 49) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CHD8_HUMAN
|
||||||
| NC score | 0.012467 (rank : 50) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 235 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCK8, Q68DQ0, Q8N3Z9, Q8NCY4, Q8TBR9, Q96F26 | Gene names | CHD8, HELSNF1, KIAA1564 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 8 (EC 3.6.1.-) (ATP- dependent helicase CHD8) (CHD-8) (Helicase with SNF2 domain 1). | |||||
|
UBP31_HUMAN
|
||||||
| NC score | 0.012129 (rank : 51) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q70CQ4, Q6AW97, Q6ZTC0, Q6ZTN2, Q9ULL7 | Gene names | USP31, KIAA1203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 31 (EC 3.1.2.15) (Ubiquitin thioesterase 31) (Ubiquitin-specific-processing protease 31) (Deubiquitinating enzyme 31). | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.011989 (rank : 52) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.011942 (rank : 53) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
TTBK2_HUMAN
|
||||||
| NC score | 0.011615 (rank : 54) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 822 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ55, O94932, Q6ZN52, Q8IVV1 | Gene names | TTBK2, KIAA0847 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tau-tubulin kinase 2 (EC 2.7.11.1). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.009568 (rank : 55) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
KIF17_MOUSE
|
||||||
| NC score | 0.009407 (rank : 56) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PW8 | Gene names | Kif17 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF17 (MmKIF17). | |||||
|
ZFY2_MOUSE
|
||||||
| NC score | 0.003963 (rank : 57) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 795 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20662 | Gene names | Zfy2, Zfy-2 | |||
|
Domain Architecture |
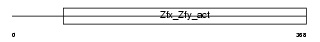
|
|||||
| Description | Zinc finger Y-chromosomal protein 2. | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.001163 (rank : 58) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 56 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||