Please be patient as the page loads
|
JADE3_HUMAN
|
||||||
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
JADE1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.979615 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978886 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.993938 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE2_HUMAN
|
||||||
| θ value | 7.25522e-167 (rank : 5) | NC score | 0.968188 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 6.7907e-165 (rank : 6) | NC score | 0.918054 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
BRPF1_HUMAN
|
||||||
| θ value | 1.19655e-52 (rank : 7) | NC score | 0.820173 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRD1_HUMAN
|
||||||
| θ value | 7.75577e-52 (rank : 8) | NC score | 0.812887 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF3_HUMAN
|
||||||
| θ value | 8.02596e-49 (rank : 9) | NC score | 0.804820 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
AF10_MOUSE
|
||||||
| θ value | 1.79631e-32 (rank : 10) | NC score | 0.800883 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF17_HUMAN
|
||||||
| θ value | 4.00176e-32 (rank : 11) | NC score | 0.835347 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
AF10_HUMAN
|
||||||
| θ value | 8.91499e-32 (rank : 12) | NC score | 0.798897 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 3.17079e-29 (rank : 13) | NC score | 0.758128 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_MOUSE
|
||||||
| θ value | 3.17079e-29 (rank : 14) | NC score | 0.771551 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
JHD3B_HUMAN
|
||||||
| θ value | 2.87452e-22 (rank : 15) | NC score | 0.612830 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 1.09232e-21 (rank : 16) | NC score | 0.608137 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3C_HUMAN
|
||||||
| θ value | 1.42661e-21 (rank : 17) | NC score | 0.595715 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
JHD3C_MOUSE
|
||||||
| θ value | 5.42112e-21 (rank : 18) | NC score | 0.599755 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3A_MOUSE
|
||||||
| θ value | 2.97466e-19 (rank : 19) | NC score | 0.599142 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 20) | NC score | 0.596694 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 21) | NC score | 0.391991 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
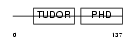
PHF1_MOUSE
|
||||||
| θ value | 3.41135e-07 (rank : 22) | NC score | 0.392168 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 23) | NC score | 0.290998 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
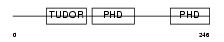
MTF2_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 24) | NC score | 0.387324 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |
|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 25) | NC score | 0.390962 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
PHF10_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 26) | NC score | 0.369113 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| θ value | 0.000121331 (rank : 27) | NC score | 0.366385 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
CHD5_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 28) | NC score | 0.186557 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
BAZ2B_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 29) | NC score | 0.244122 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
CHD4_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 30) | NC score | 0.181728 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD4_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 31) | NC score | 0.180173 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 32) | NC score | 0.245941 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NSD1_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 33) | NC score | 0.237710 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
NSD1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 34) | NC score | 0.241278 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 35) | NC score | 0.219610 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 36) | NC score | 0.213751 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 37) | NC score | 0.258940 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 38) | NC score | 0.276267 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
PHF6_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.231308 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
CHD3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 40) | NC score | 0.181465 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 41) | NC score | 0.257330 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 42) | NC score | 0.207280 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 43) | NC score | 0.202723 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
PHF6_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 44) | NC score | 0.213359 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 45) | NC score | 0.202971 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
AIRE_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 46) | NC score | 0.325194 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
DPF1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 47) | NC score | 0.254921 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 48) | NC score | 0.237679 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
JAD1D_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 49) | NC score | 0.295838 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
PHF7_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 50) | NC score | 0.279927 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
REQU_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 51) | NC score | 0.249462 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 52) | NC score | 0.218101 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 53) | NC score | 0.216842 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
TIF1G_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 54) | NC score | 0.195619 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 55) | NC score | 0.235213 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
MYST4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 56) | NC score | 0.223570 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
REQU_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 57) | NC score | 0.244178 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 58) | NC score | 0.214435 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 59) | NC score | 0.244450 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
K1333_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 60) | NC score | 0.189627 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
TIF1G_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 61) | NC score | 0.192592 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 62) | NC score | 0.314185 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
BAZ1A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 63) | NC score | 0.246985 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 64) | NC score | 0.226255 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PHF7_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 65) | NC score | 0.246108 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MEOX1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 66) | NC score | 0.020124 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50221 | Gene names | MEOX1, MOX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-1 (Mesenchyme homeobox 1). | |||||
|
PHF11_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 67) | NC score | 0.245830 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
PHF12_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 68) | NC score | 0.236737 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 69) | NC score | 0.234580 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 70) | NC score | 0.196887 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 71) | NC score | 0.267980 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 72) | NC score | 0.243761 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 73) | NC score | 0.198115 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TRI66_HUMAN
|
||||||
| θ value | 0.125558 (rank : 74) | NC score | 0.209491 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
UHRF2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 75) | NC score | 0.194813 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
DPF1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 76) | NC score | 0.244012 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
JAD1C_HUMAN
|
||||||
| θ value | 0.163984 (rank : 77) | NC score | 0.269509 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| θ value | 0.163984 (rank : 78) | NC score | 0.269432 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
TRI66_MOUSE
|
||||||
| θ value | 0.163984 (rank : 79) | NC score | 0.209953 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
UHRF2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 80) | NC score | 0.176500 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
JAD1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 81) | NC score | 0.276470 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 82) | NC score | 0.197507 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TIF1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 83) | NC score | 0.200725 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
MEOX1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 84) | NC score | 0.020832 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32442 | Gene names | Meox1, Mox-1, Mox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-1 (Mesenchyme homeobox 1). | |||||
|
TFAP4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 85) | NC score | 0.027906 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 0.365318 (rank : 86) | NC score | 0.199271 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
XE7_HUMAN
|
||||||
| θ value | 0.365318 (rank : 87) | NC score | 0.018768 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
PF21A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 88) | NC score | 0.232525 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
ING3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 89) | NC score | 0.121357 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
PF21A_MOUSE
|
||||||
| θ value | 0.813845 (rank : 90) | NC score | 0.230975 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
TCF20_HUMAN
|
||||||
| θ value | 0.813845 (rank : 91) | NC score | 0.144319 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 0.813845 (rank : 92) | NC score | 0.141184 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 1.06291 (rank : 93) | NC score | 0.012535 (rank : 168) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
EPC1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 94) | NC score | 0.142619 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2F5, Q5VW54, Q5VW56, Q5VW58, Q8NAQ4, Q8NE21, Q96LF4, Q96RR6, Q9H7T7 | Gene names | EPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
EPC1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 95) | NC score | 0.131694 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C9X6, Q9Z299 | Gene names | Epc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
EPC2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 96) | NC score | 0.123953 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
EPC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 97) | NC score | 0.124243 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C0I4, Q3URX9, Q80Y70, Q8BXC5 | Gene names | Epc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
ING3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 98) | NC score | 0.117468 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
LRC41_HUMAN
|
||||||
| θ value | 1.06291 (rank : 99) | NC score | 0.033017 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15345, Q71RA8, Q9BSM0 | Gene names | LRRC41, MUF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
LRC41_MOUSE
|
||||||
| θ value | 1.06291 (rank : 100) | NC score | 0.030570 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1C9, Q91VX4 | Gene names | Lrrc41, Muf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
LRRK2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 101) | NC score | -0.002332 (rank : 189) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||
|
PHF22_MOUSE
|
||||||
| θ value | 1.06291 (rank : 102) | NC score | 0.146200 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 103) | NC score | 0.028155 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 104) | NC score | 0.021726 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
PHF20_HUMAN
|
||||||
| θ value | 1.38821 (rank : 105) | NC score | 0.082701 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 1.38821 (rank : 106) | NC score | 0.085956 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PLCB3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 107) | NC score | 0.012529 (rank : 169) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 108) | NC score | 0.059539 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
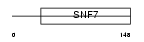
CHM4A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 109) | NC score | 0.013645 (rank : 166) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |
|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
CJ082_HUMAN
|
||||||
| θ value | 2.36792 (rank : 110) | NC score | 0.029330 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WW14 | Gene names | C10orf82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf82. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 111) | NC score | 0.008546 (rank : 175) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 112) | NC score | 0.023108 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 113) | NC score | 0.005435 (rank : 179) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 4.03905 (rank : 114) | NC score | 0.020767 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 4.03905 (rank : 115) | NC score | 0.038588 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 4.03905 (rank : 116) | NC score | 0.019740 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
TB182_HUMAN
|
||||||
| θ value | 4.03905 (rank : 117) | NC score | 0.023654 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
UHRF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 118) | NC score | 0.152064 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | -0.001689 (rank : 188) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
ING4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.091264 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.090132 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 122) | NC score | 0.010689 (rank : 172) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 5.27518 (rank : 123) | NC score | -0.000126 (rank : 187) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
SGOL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 124) | NC score | 0.012776 (rank : 167) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TSY8, Q811I4, Q8C9C4, Q8CGJ4, Q9CS55 | Gene names | Sgol2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2. | |||||
|
UHRF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 125) | NC score | 0.149738 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
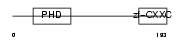
CXCC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.069998 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |
|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.068119 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXXC6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.021270 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.007362 (rank : 176) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | 0.016793 (rank : 164) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.006408 (rank : 177) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
K1333_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.131983 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
PLCB3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.011088 (rank : 171) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
POK9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.004894 (rank : 181) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63134 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pol protein (HERV-K104 Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49)]. | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.005712 (rank : 178) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.011492 (rank : 170) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TRI42_HUMAN
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.021858 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
TSP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.003855 (rank : 183) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
UBP16_HUMAN
|
||||||
| θ value | 6.88961 (rank : 139) | NC score | 0.004727 (rank : 182) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.020023 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
GCAB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.001091 (rank : 186) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01864 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2A chain C region secreted form (B allele). | |||||
|
GLE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.013813 (rank : 165) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
PHF22_HUMAN
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.094656 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
POK12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.002173 (rank : 185) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63135 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pol protein (HERV-K102 Pol protein) (HERV-K(III) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
SETBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.010664 (rank : 173) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
TBC12_HUMAN
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.004969 (rank : 180) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
UBE3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.002222 (rank : 184) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
URFB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.010021 (rank : 174) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
BRD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.074279 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
BRD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.067549 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.071817 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.067808 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.075365 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRD7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.107397 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.104713 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
BRD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.082347 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
BRD8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.079735 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
BRD9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.110041 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.108369 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRDT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.062217 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.062373 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
CBP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.065928 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CBP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.060521 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.084165 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
DPF3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.053846 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58269 | Gene names | Dpf3, Cerd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-finger protein Dpf3 (cer-d4). | |||||
|
EP300_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.063508 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
GCNL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.085004 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.084507 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
ING1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.056675 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UK53, O00532, O43658, Q9H007, Q9HD98, Q9HD99, Q9NS83, Q9P0U6, Q9UBC6, Q9UIJ1, Q9UIJ2, Q9UIJ3, Q9UIJ4, Q9UK52 | Gene names | ING1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.055898 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QXV3, Q9QUP8, Q9QXV4, Q9QZX3 | Gene names | Ing1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.059627 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WYH8, Q9BS30 | Gene names | ING5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5 (p28ING5). | |||||
|
ING5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.058152 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
JARD2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.117353 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JARD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.118456 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
JHD3D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.239550 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.240811 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
LY10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.132510 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
PB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.072363 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
PCAF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.083885 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
PF21B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.142283 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
PF21B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.138670 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.053792 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.057168 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.061368 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SP110_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.088509 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
TAF1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.076072 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
TAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.078197 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.080957 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZMY11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.091777 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
JADE3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 148 | |
| SwissProt Accessions | Q92613, Q6IE79 | Gene names | PHF16, JADE3, KIAA0215 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE3_MOUSE
|
||||||
| NC score | 0.993938 (rank : 2) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q6IE82, Q6ZQG2 | Gene names | Phf16, Jade3, Kiaa0215 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-3 (PHD finger protein 16). | |||||
|
JADE1_HUMAN
|
||||||
| NC score | 0.979615 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q6IE81, Q4W5D5, Q6ZSL7, Q8NC41, Q96JL8, Q96SQ1, Q9H692 | Gene names | PHF17, JADE1, KIAA1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE1_MOUSE
|
||||||
| NC score | 0.978886 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q6ZPI0, Q6IE84, Q8C7S6, Q8CFM2, Q8R362 | Gene names | Phf17, Jade1, Kiaa1807 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-1 (PHD finger protein 17). | |||||
|
JADE2_HUMAN
|
||||||
| NC score | 0.968188 (rank : 5) | θ value | 7.25522e-167 (rank : 5) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q9NQC1, Q6IE80, Q8TEK0, Q92513, Q96GQ6 | Gene names | PHF15, JADE2, KIAA0239 | |||
|
Domain Architecture |

|
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.918054 (rank : 6) | θ value | 6.7907e-165 (rank : 6) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
AF17_HUMAN
|
||||||
| NC score | 0.835347 (rank : 7) | θ value | 4.00176e-32 (rank : 11) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P55198, Q9UF49 | Gene names | MLLT6, AF17 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-17. | |||||
|
BRPF1_HUMAN
|
||||||
| NC score | 0.820173 (rank : 8) | θ value | 1.19655e-52 (rank : 7) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | P55201, Q9UHI0 | Gene names | BRPF1, BR140 | |||
|
Domain Architecture |

|
|||||
| Description | Peregrin (Bromodomain and PHD finger-containing protein 1) (BR140 protein). | |||||
|
BRD1_HUMAN
|
||||||
| NC score | 0.812887 (rank : 9) | θ value | 7.75577e-52 (rank : 8) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O95696 | Gene names | BRD1, BRL, BRPF2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 1 (BR140-like protein). | |||||
|
BRPF3_HUMAN
|
||||||
| NC score | 0.804820 (rank : 10) | θ value | 8.02596e-49 (rank : 9) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9ULD4, Q5R3K8 | Gene names | BRPF3, KIAA1286 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain and PHD finger-containing protein 3. | |||||
|
AF10_MOUSE
|
||||||
| NC score | 0.800883 (rank : 11) | θ value | 1.79631e-32 (rank : 10) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | O54826 | Gene names | Mllt10, Af10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
AF10_HUMAN
|
||||||
| NC score | 0.798897 (rank : 12) | θ value | 8.91499e-32 (rank : 12) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P55197 | Gene names | MLLT10, AF10 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-10. | |||||
|
PHF14_MOUSE
|
||||||
| NC score | 0.771551 (rank : 13) | θ value | 3.17079e-29 (rank : 14) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9D4H9, Q810Z6, Q8C284, Q8CGF8, Q9D1Q8 | Gene names | Phf14 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 14. | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.758128 (rank : 14) | θ value | 3.17079e-29 (rank : 13) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
JHD3B_HUMAN
|
||||||
| NC score | 0.612830 (rank : 15) | θ value | 2.87452e-22 (rank : 15) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O94953, O75274, Q6P3R5, Q9P1V1, Q9UF40 | Gene names | JMJD2B, JHDM3B, KIAA0876 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.608137 (rank : 16) | θ value | 1.09232e-21 (rank : 16) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
JHD3C_MOUSE
|
||||||
| NC score | 0.599755 (rank : 17) | θ value | 5.42112e-21 (rank : 18) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8VCD7, Q3UNP7, Q69ZZ5, Q8BUY6, Q8BWA1 | Gene names | Jmjd2c, Jhdm3c, Kiaa0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C). | |||||
|
JHD3A_MOUSE
|
||||||
| NC score | 0.599142 (rank : 18) | θ value | 2.97466e-19 (rank : 19) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q8BW72, Q3UKM5, Q3UM81, Q3UWV2, Q6ZQ72, Q8K137 | Gene names | Jmjd2a, Jhdm3a, Jmjd2, Kiaa0677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3A_HUMAN
|
||||||
| NC score | 0.596694 (rank : 19) | θ value | 6.62687e-19 (rank : 20) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75164, Q5VVB1 | Gene names | JMJD2A, JHDM3A, JMJD2, KIAA0677 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3A (EC 1.14.11.-) (Jumonji domain-containing protein 2A). | |||||
|
JHD3C_HUMAN
|
||||||
| NC score | 0.595715 (rank : 20) | θ value | 1.42661e-21 (rank : 17) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9H3R0, O94877, Q2M3M0, Q5JUC9, Q5VYJ2, Q5VYJ3 | Gene names | JMJD2C, GASC1, JHDM3C, KIAA0780 | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3C (EC 1.14.11.-) (Jumonji domain-containing protein 2C) (Gene amplified in squamous cell carcinoma 1 protein) (GASC-1 protein). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.392168 (rank : 21) | θ value | 3.41135e-07 (rank : 22) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.391991 (rank : 22) | θ value | 3.41135e-07 (rank : 21) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
MTF2_MOUSE
|
||||||
| NC score | 0.390962 (rank : 23) | θ value | 1.69304e-06 (rank : 25) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q02395, Q569Z8, Q6PG89, Q8BGP9, Q8BSJ7 | Gene names | Mtf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-response element-binding transcription factor 2 (Zinc-regulated factor 1) (ZiRF1) (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
MTF2_HUMAN
|
||||||
| NC score | 0.387324 (rank : 24) | θ value | 1.69304e-06 (rank : 24) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9Y483, Q9UES9, Q9UP40 | Gene names | MTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Metal-response element-binding transcription factor 2 (Metal-response element DNA-binding protein M96) (Metal-regulatory transcription factor 2) (PCL2). | |||||
|
PHF10_HUMAN
|
||||||
| NC score | 0.369113 (rank : 25) | θ value | 9.29e-05 (rank : 26) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8WUB8, Q2YDA3, Q9BXD2, Q9NV26 | Gene names | PHF10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10 (XAP135). | |||||
|
PHF10_MOUSE
|
||||||
| NC score | 0.366385 (rank : 26) | θ value | 0.000121331 (rank : 27) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q9D8M7, Q99LV5 | Gene names | Phf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 10. | |||||
|
AIRE_HUMAN
|
||||||
| NC score | 0.325194 (rank : 27) | θ value | 0.00665767 (rank : 46) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | O43918, O43922, O43932, O75745 | Gene names | AIRE, APECED | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein) (APECED protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.314185 (rank : 28) | θ value | 0.0563607 (rank : 62) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
JAD1D_HUMAN
|
||||||
| NC score | 0.295838 (rank : 29) | θ value | 0.00869519 (rank : 49) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | Q9BY66, Q92509, Q92809, Q9HCU1 | Gene names | SMCY, HY, HYA, JARID1D, KIAA0234 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.290998 (rank : 30) | θ value | 1.69304e-06 (rank : 23) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
PHF7_MOUSE
|
||||||
| NC score | 0.279927 (rank : 31) | θ value | 0.00869519 (rank : 50) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9DAG9, Q6PG81 | Gene names | Phf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
JAD1A_HUMAN
|
||||||
| NC score | 0.276470 (rank : 32) | θ value | 0.279714 (rank : 81) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | P29375 | Gene names | JARID1A, RBBP2, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1A (Retinoblastoma-binding protein 2) (RBBP-2). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.276267 (rank : 33) | θ value | 0.00175202 (rank : 38) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
JAD1C_HUMAN
|
||||||
| NC score | 0.269509 (rank : 34) | θ value | 0.163984 (rank : 77) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | P41229 | Gene names | SMCX, DXS1272E, JARID1C, XE169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (Protein SmcX) (Protein Xe169). | |||||
|
JAD1C_MOUSE
|
||||||
| NC score | 0.269432 (rank : 35) | θ value | 0.163984 (rank : 78) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P41230, O54995, Q9CVI4, Q9D0C3, Q9QVR8, Q9R039 | Gene names | Smcx, Jarid1c, Xe169 | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1C (SmcX protein) (Xe169 protein). | |||||
|
JAD1D_MOUSE
|
||||||
| NC score | 0.267980 (rank : 36) | θ value | 0.0961366 (rank : 71) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q62240, Q9QVR9, Q9R040 | Gene names | Smcy, Hya, Jarid1d | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji/ARID domain-containing protein 1D (SmcY protein) (Histocompatibility Y antigen) (H-Y). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.258940 (rank : 37) | θ value | 0.00175202 (rank : 37) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.257330 (rank : 38) | θ value | 0.00228821 (rank : 41) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
DPF1_MOUSE
|
||||||
| NC score | 0.254921 (rank : 39) | θ value | 0.00665767 (rank : 47) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9QX66, Q9QX65, Q9QYA4 | Gene names | Dpf1, Neud4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
REQU_HUMAN
|
||||||
| NC score | 0.249462 (rank : 40) | θ value | 0.0113563 (rank : 51) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 523 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q92785 | Gene names | DPF2, REQ, UBID4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
BAZ1A_HUMAN
|
||||||
| NC score | 0.246985 (rank : 41) | θ value | 0.0563607 (rank : 63) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q9NRL2, Q9NZ15, Q9P065, Q9UIG1, Q9Y3V3 | Gene names | BAZ1A, ACF1, WCRF180 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1A (ATP-utilizing chromatin assembly and remodeling factor 1) (hACF1) (ATP-dependent chromatin remodelling protein) (Williams syndrome transcription factor-related chromatin remodeling factor 180) (WCRF180) (hWALp1) (CHRAC subunit ACF1). | |||||
|
PHF7_HUMAN
|
||||||
| NC score | 0.246108 (rank : 42) | θ value | 0.0563607 (rank : 65) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BWX1 | Gene names | PHF7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 7 (Testis development protein NYD-SP6). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.245941 (rank : 43) | θ value | 0.00134147 (rank : 32) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PHF11_HUMAN
|
||||||
| NC score | 0.245830 (rank : 44) | θ value | 0.0736092 (rank : 67) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UIL8, Q9Y5A2 | Gene names | PHF11, BCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 11 (BRCA1 C-terminus-associated protein) (NY-REN-34 antigen). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.244450 (rank : 45) | θ value | 0.0252991 (rank : 59) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
REQU_MOUSE
|
||||||
| NC score | 0.244178 (rank : 46) | θ value | 0.0193708 (rank : 57) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q61103, Q3UNP5, Q60663, Q9QYA3 | Gene names | Dpf2, Req, Ubid4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein ubi-d4 (Requiem) (Apoptosis response zinc finger protein) (D4, zinc and double PHD fingers family 2). | |||||
|
BAZ2B_HUMAN
|
||||||
| NC score | 0.244122 (rank : 47) | θ value | 0.00102713 (rank : 29) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q9UIF8, Q96EA1, Q96SQ8, Q9P252, Q9Y4N8 | Gene names | BAZ2B, KIAA1476 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2B (hWALp4). | |||||
|
DPF1_HUMAN
|
||||||
| NC score | 0.244012 (rank : 48) | θ value | 0.163984 (rank : 76) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q92782 | Gene names | DPF1, NEUD4 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc-finger protein neuro-d4 (D4, zinc and double PHD fingers family 1). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.243761 (rank : 49) | θ value | 0.0961366 (rank : 72) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
NSD1_MOUSE
|
||||||
| NC score | 0.241278 (rank : 50) | θ value | 0.00134147 (rank : 34) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 472 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | O88491, Q8C480, Q9CT70 | Gene names | Nsd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein). | |||||
|
JHD3D_MOUSE
|
||||||
| NC score | 0.240811 (rank : 51) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q3U2K5, Q2M1G7, Q8BI19 | Gene names | Jmjd2d, Jhdm3d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
JHD3D_HUMAN
|
||||||
| NC score | 0.239550 (rank : 52) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6B0I6, Q9NT41, Q9NW76 | Gene names | JMJD2D, JHDM3D | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 3D (EC 1.14.11.-) (Jumonji domain-containing protein 2D). | |||||
|
NSD1_HUMAN
|
||||||
| NC score | 0.237710 (rank : 53) | θ value | 0.00134147 (rank : 33) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96L73, Q96PD8, Q96RN7 | Gene names | NSD1, ARA267 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific (EC 2.1.1.43) (H3-K36-HMTase) (H4-K20-HMTase) (Nuclear receptor-binding SET domain-containing protein 1) (NR-binding SET domain-containing protein) (Androgen receptor-associated coregulator 267). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.237679 (rank : 54) | θ value | 0.00869519 (rank : 48) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.236737 (rank : 55) | θ value | 0.0736092 (rank : 68) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.235213 (rank : 56) | θ value | 0.0193708 (rank : 55) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.234580 (rank : 57) | θ value | 0.0736092 (rank : 69) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
PF21A_HUMAN
|
||||||
| NC score | 0.232525 (rank : 58) | θ value | 0.62314 (rank : 88) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96BD5, Q6AWA2, Q9C0G7, Q9H8V9, Q9HAK6, Q9NZE9 | Gene names | PHF21A, BHC80, KIAA1696 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (BHC80a). | |||||
|
PHF6_HUMAN
|
||||||
| NC score | 0.231308 (rank : 59) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWS0, Q96JK3, Q9BRU0 | Gene names | PHF6, KIAA1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6 (PHD-like zinc finger protein). | |||||
|
PF21A_MOUSE
|
||||||
| NC score | 0.230975 (rank : 60) | θ value | 0.813845 (rank : 90) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6ZPK0, Q6XVG0, Q80Z33, Q8CAZ4, Q8VEC8 | Gene names | Phf21a, Bhc80, Kiaa1696, Pftf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21A (BRAF35-HDAC complex protein BHC80) (mBHC80) (BHC80a). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.226255 (rank : 61) | θ value | 0.0563607 (rank : 64) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MYST4_MOUSE
|
||||||
| NC score | 0.223570 (rank : 62) | θ value | 0.0193708 (rank : 56) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q8BRB7, Q7TNW5, Q8BG35, Q8C441, Q9JKX5 | Gene names | Myst4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Querkopf protein). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.219610 (rank : 63) | θ value | 0.00134147 (rank : 35) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.218101 (rank : 64) | θ value | 0.0148317 (rank : 52) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.216842 (rank : 65) | θ value | 0.0148317 (rank : 53) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.214435 (rank : 66) | θ value | 0.0252991 (rank : 58) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.213751 (rank : 67) | θ value | 0.00134147 (rank : 36) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
PHF6_MOUSE
|
||||||
| NC score | 0.213359 (rank : 68) | θ value | 0.00390308 (rank : 44) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D4J7, Q80T86, Q8BYT8, Q8C1B5 | Gene names | Phf6, Kiaa1823 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 6. | |||||
|
TRI66_MOUSE
|
||||||
| NC score | 0.209953 (rank : 69) | θ value | 0.163984 (rank : 79) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q924W6 | Gene names | Trim66 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TRI66_HUMAN
|
||||||
| NC score | 0.209491 (rank : 70) | θ value | 0.125558 (rank : 74) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | O15016, Q9BQQ4 | Gene names | TRIM66, KIAA0298 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 66. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.207280 (rank : 71) | θ value | 0.00228821 (rank : 42) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.202971 (rank : 72) | θ value | 0.00509761 (rank : 45) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.202723 (rank : 73) | θ value | 0.00228821 (rank : 43) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TIF1A_HUMAN
|
||||||
| NC score | 0.200725 (rank : 74) | θ value | 0.279714 (rank : 83) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | O15164, O95854 | Gene names | TRIM24, RNF82, TIF1, TIF1A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24) (RING finger protein 82). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.199271 (rank : 75) | θ value | 0.365318 (rank : 86) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.198115 (rank : 76) | θ value | 0.125558 (rank : 73) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.197507 (rank : 77) | θ value | 0.279714 (rank : 82) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.196887 (rank : 78) | θ value | 0.0961366 (rank : 70) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
TIF1G_MOUSE
|
||||||
| NC score | 0.195619 (rank : 79) | θ value | 0.0148317 (rank : 54) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q99PP7, Q6SI71, Q6ZPX5 | Gene names | Trim33, Kiaa1113 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33). | |||||
|
UHRF2_MOUSE
|
||||||
| NC score | 0.194813 (rank : 80) | θ value | 0.125558 (rank : 75) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q7TMI3, Q8BG56, Q8BJP6, Q8BY30, Q8K1I5 | Gene names | Uhrf2, Nirf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95-like ring finger protein) (Nuclear zinc finger protein Np97) (NIRF). | |||||
|
TIF1G_HUMAN
|
||||||
| NC score | 0.192592 (rank : 81) | θ value | 0.0330416 (rank : 61) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q9UPN9, O95855, Q9C017, Q9UJ79 | Gene names | TRIM33, KIAA1113, RFG7, TIF1G | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-gamma (TIF1-gamma) (Tripartite motif-containing protein 33) (RET-fused gene 7 protein) (Rfg7 protein). | |||||
|
K1333_MOUSE
|
||||||
| NC score | 0.189627 (rank : 82) | θ value | 0.0330416 (rank : 60) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q5RJY2, Q6ZPT7, Q8BNA4 | Gene names | Kiaa1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
CHD5_HUMAN
|
||||||
| NC score | 0.186557 (rank : 83) | θ value | 0.00035302 (rank : 28) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8TDI0, O75032, Q5TG89, Q9UFR9 | Gene names | CHD5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 5 (EC 3.6.1.-) (ATP- dependent helicase CHD5) (CHD-5). | |||||
|
CHD4_HUMAN
|
||||||
| NC score | 0.181728 (rank : 84) | θ value | 0.00134147 (rank : 30) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q14839, Q8IXZ5 | Gene names | CHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (EC 3.6.1.-) (ATP- dependent helicase CHD4) (CHD-4) (Mi-2 autoantigen 218 kDa protein) (Mi2-beta). | |||||
|
CHD3_HUMAN
|
||||||
| NC score | 0.181465 (rank : 85) | θ value | 0.00228821 (rank : 40) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | Q12873, Q9Y4I0 | Gene names | CHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain helicase-DNA-binding protein 3 (EC 3.6.1.-) (ATP- dependent helicase CHD3) (CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha) (Zinc-finger helicase) (hZFH). | |||||
|
CHD4_MOUSE
|
||||||
| NC score | 0.180173 (rank : 86) | θ value | 0.00134147 (rank : 31) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q6PDQ2 | Gene names | Chd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain helicase-DNA-binding protein 4 (CHD-4). | |||||
|
UHRF2_HUMAN
|
||||||
| NC score | 0.176500 (rank : 87) | θ value | 0.21417 (rank : 80) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96PU4, Q5VYR1, Q5VYR3, Q659C8, Q8TAG7 | Gene names | UHRF2, NIRF, RNF107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 2 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 2) (Np95/ICBP90-like RING finger protein) (Np95-like RING finger protein) (Nuclear zinc finger protein Np97) (RING finger protein 107). | |||||
|
UHRF1_HUMAN
|
||||||
| NC score | 0.152064 (rank : 88) | θ value | 4.03905 (rank : 118) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q96T88, Q8J022, Q9H6S6, Q9P115, Q9P1U7 | Gene names | UHRF1, ICBP90, NP95, RNF106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Inverted CCAAT box-binding protein of 90 kDa) (Transcription factor ICBP90) (Nuclear zinc finger protein Np95) (Nuclear protein 95) (HuNp95) (RING finger protein 106). | |||||
|
UHRF1_MOUSE
|
||||||
| NC score | 0.149738 (rank : 89) | θ value | 5.27518 (rank : 125) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8VDF2, Q8C6F1, Q8VIA1, Q9Z1H6 | Gene names | Uhrf1, Np95 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-like PHD and RING finger domain-containing protein 1 (EC 6.3.2.-) (Ubiquitin-like-containing PHD and RING finger domains protein 1) (Nuclear zinc finger protein Np95) (Nuclear protein 95). | |||||
|
PHF22_MOUSE
|
||||||
| NC score | 0.146200 (rank : 90) | θ value | 1.06291 (rank : 102) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D168, Q3U2Q5, Q921U2 | Gene names | Ints12, Phf22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
TCF20_HUMAN
|
||||||
| NC score | 0.144319 (rank : 91) | θ value | 0.813845 (rank : 91) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9UGU0, O14528, Q13078, Q4V353, Q9H4M0 | Gene names | TCF20, KIAA0292, SPBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP) (AR1). | |||||
|
EPC1_HUMAN
|
||||||
| NC score | 0.142619 (rank : 92) | θ value | 1.06291 (rank : 94) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H2F5, Q5VW54, Q5VW56, Q5VW58, Q8NAQ4, Q8NE21, Q96LF4, Q96RR6, Q9H7T7 | Gene names | EPC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
PF21B_HUMAN
|
||||||
| NC score | 0.142283 (rank : 93) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q96EK2, Q5TFL2, Q6ICC0 | Gene names | PHF21B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.141184 (rank : 94) | θ value | 0.813845 (rank : 92) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
PF21B_MOUSE
|
||||||
| NC score | 0.138670 (rank : 95) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8C966 | Gene names | Phf21b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 21B. | |||||
|
LY10_HUMAN
|
||||||
| NC score | 0.132510 (rank : 96) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q13342, Q13341, Q92881, Q96TG3 | Gene names | SP140, LYSP100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear body protein SP140 (Nuclear autoantigen Sp-140) (Speckled 140 kDa) (LYSp100 protein) (Lymphoid-restricted homolog of Sp100). | |||||
|
K1333_HUMAN
|
||||||
| NC score | 0.131983 (rank : 97) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7L622, Q9BVR2, Q9H9E9, Q9NXC0, Q9P2L3 | Gene names | KIAA1333 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1333 (EC 6.3.2.-). | |||||
|
EPC1_MOUSE
|
||||||
| NC score | 0.131694 (rank : 98) | θ value | 1.06291 (rank : 95) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8C9X6, Q9Z299 | Gene names | Epc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 1. | |||||
|
EPC2_MOUSE
|
||||||
| NC score | 0.124243 (rank : 99) | θ value | 1.06291 (rank : 97) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8C0I4, Q3URX9, Q80Y70, Q8BXC5 | Gene names | Epc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
EPC2_HUMAN
|
||||||
| NC score | 0.123953 (rank : 100) | θ value | 1.06291 (rank : 96) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q52LR7, Q7L9J1, Q96RR7, Q9NUT8, Q9NVR1, Q9UFM9 | Gene names | EPC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enhancer of polycomb homolog 2. | |||||
|
ING3_MOUSE
|
||||||
| NC score | 0.121357 (rank : 101) | θ value | 0.813845 (rank : 89) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8VEK6, Q99JS6, Q9ERB2 | Gene names | Ing3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.118456 (rank : 102) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
ING3_HUMAN
|
||||||
| NC score | 0.117468 (rank : 103) | θ value | 1.06291 (rank : 98) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9NXR8, O60394, Q6GMT3, Q7Z762, Q969G0, Q96DT4, Q9HC99, Q9P081 | Gene names | ING3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 3 (p47ING3 protein). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.117353 (rank : 104) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
BRD9_HUMAN
|
||||||
| NC score | 0.110041 (rank : 105) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H8M2, Q2XUS1, Q6UWU9, Q8IUS4, Q9H5Q5, Q9H7R9 | Gene names | BRD9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9 (Rhabdomyosarcoma antigen MU-RMS- 40.8). | |||||
|
BRD9_MOUSE
|
||||||
| NC score | 0.108369 (rank : 106) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3UQU0, Q5FWH1, Q811F7 | Gene names | Brd9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 9. | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.107397 (rank : 107) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.104713 (rank : 108) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
PHF22_HUMAN
|
||||||
| NC score | 0.094656 (rank : 109) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
ZMY11_MOUSE
|
||||||
| NC score | 0.091777 (rank : 110) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8R5C8, Q8C155 | Gene names | Zmynd11 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11. | |||||
|
ING4_HUMAN
|
||||||
| NC score | 0.091264 (rank : 111) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9UNL4, Q96E15, Q9H3J0 | Gene names | ING4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
ING4_MOUSE
|
||||||
| NC score | 0.090132 (rank : 112) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8C0D7, Q8C1S7, Q8K3Q5, Q8K3Q6, Q8K3Q7, Q9D7F9 | Gene names | Ing4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 4 (p29ING4). | |||||
|
SP110_HUMAN
|
||||||
| NC score | 0.088509 (rank : 113) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9HB58, Q14976, Q14977, Q8WUZ6, Q9HCT8 | Gene names | SP110 | |||
|
Domain Architecture |

|
|||||
| Description | Sp110 nuclear body protein (Speckled 110 kDa) (Transcriptional coactivator Sp110) (Interferon-induced protein 41/75). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.085956 (rank : 114) | θ value | 1.38821 (rank : 106) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
GCNL2_HUMAN
|
||||||
| NC score | 0.085004 (rank : 115) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q92830, Q9UCW1 | Gene names | GCN5L2, GCN5, HGCN5 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (hsGCN5) (STAF97). | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.084507 (rank : 116) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.084165 (rank : 117) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
PCAF_HUMAN
|
||||||
| NC score | 0.083885 (rank : 118) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q92831, Q6NSK1 | Gene names | PCAF | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase PCAF (EC 2.3.1.48) (P300/CBP-associated factor) (P/CAF) (Histone acetylase PCAF). | |||||
|
PHF20_HUMAN
|
||||||
| NC score | 0.082701 (rank : 119) | θ value | 1.38821 (rank : 105) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BVI0, Q5JWY9, Q9BWV4, Q9BXA3, Q9BZW3, Q9H421, Q9H4J6, Q9NZ22 | Gene names | PHF20, C20orf104, GLEA2, HCA58, NZF, TZP | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58) (Glioma-expressed antigen 2) (Transcription factor TZP) (Novel zinc finger protein). | |||||
|
BRD8_HUMAN
|
||||||
| NC score | 0.082347 (rank : 120) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9H0E9, O43178, Q15355, Q59GN0, Q969M9 | Gene names | BRD8, SMAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8 (p120) (Skeletal muscle abundant protein) (Thyroid hormone receptor coactivating protein 120kDa) (TrCP120). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.080957 (rank : 121) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
BRD8_MOUSE
|
||||||
| NC score | 0.079735 (rank : 122) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R3B7, Q8C049, Q8R583, Q8VDP0 | Gene names | Brd8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 8. | |||||
|
TAF1_HUMAN
|
||||||
| NC score | 0.078197 (rank : 123) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P21675, Q6IUZ1 | Gene names | TAF1, BA2R, CCG1, TAF2A | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 1 (EC 2.7.11.1) (Transcription initiation factor TFIID 250 kDa subunit) (TAF(II)250) (TAFII-250) (TAFII250) (TBP-associated factor 250 kDa) (p250) (Cell cycle gene 1 protein). | |||||
|
TAF1L_HUMAN
|
||||||
| NC score | 0.076072 (rank : 124) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IZX4 | Gene names | TAF1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID 210 kDa subunit (TBP-associated factor 210 kDa) (TAF(II)210) (TBP-associated factor 1-like). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.075365 (rank : 125) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.074279 (rank : 126) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
PB1_HUMAN
|
||||||
| NC score | 0.072363 (rank : 127) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86U86, Q9H2T3, Q9H2T4, Q9H2T5, Q9H301, Q9H314 | Gene names | PB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein polybromo-1 (hPB1) (Polybromo-1D) (BRG1-associated factor 180) (BAF180). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.071817 (rank : 128) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CXCC1_HUMAN
|
||||||
| NC score | 0.069998 (rank : 129) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9P0U4, Q8N2W4, Q96BC8, Q9P2V7 | Gene names | CXXC1, CGBP, PCCX1, PHF18 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
CXCC1_MOUSE
|
||||||
| NC score | 0.068119 (rank : 130) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9CWW7 | Gene names | Cxxc1, Cgbp, Pccx1 | |||
|
Domain Architecture |

|
|||||
| Description | CpG-binding protein (PHD finger and CXXC domain-containing protein 1) (CXXC-type zinc finger protein 1). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.067808 (rank : 131) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRD3_HUMAN
|
||||||
| NC score | 0.067549 (rank : 132) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15059, Q5T1R7, Q8N5M3, Q92645 | Gene names | BRD3, KIAA0043, RING3L | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (RING3-like protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.065928 (rank : 133) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.063508 (rank : 134) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.062373 (rank : 135) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
BRDT_HUMAN
|
||||||
| NC score | 0.062217 (rank : 136) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q58F21, O14789, Q6P5T1, Q7Z4A6, Q8IWI6 | Gene names | BRDT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.061368 (rank : 137) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.060521 (rank : 138) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ING5_HUMAN
|
||||||
| NC score | 0.059627 (rank : 139) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8WYH8, Q9BS30 | Gene names | ING5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5 (p28ING5). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.059539 (rank : 140) | θ value | 1.81305 (rank : 108) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ING5_MOUSE
|
||||||
| NC score | 0.058152 (rank : 141) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9D8Y8, Q3UL57, Q6P292, Q9CV64, Q9D9V8 | Gene names | Ing5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 5. | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.057168 (rank : 142) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ING1_HUMAN
|
||||||
| NC score | 0.056675 (rank : 143) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UK53, O00532, O43658, Q9H007, Q9HD98, Q9HD99, Q9NS83, Q9P0U6, Q9UBC6, Q9UIJ1, Q9UIJ2, Q9UIJ3, Q9UIJ4, Q9UK52 | Gene names | ING1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
ING1_MOUSE
|
||||||
| NC score | 0.055898 (rank : 144) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9QXV3, Q9QUP8, Q9QXV4, Q9QZX3 | Gene names | Ing1 | |||
|
Domain Architecture |

|
|||||
| Description | Inhibitor of growth protein 1. | |||||
|
DPF3_MOUSE
|
||||||
| NC score | 0.053846 (rank : 145) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P58269 | Gene names | Dpf3, Cerd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc-finger protein Dpf3 (cer-d4). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.053792 (rank : 146) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.038588 (rank : 147) | θ value | 4.03905 (rank : 115) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LRC41_HUMAN
|
||||||
| NC score | 0.033017 (rank : 148) | θ value | 1.06291 (rank : 99) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15345, Q71RA8, Q9BSM0 | Gene names | LRRC41, MUF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
LRC41_MOUSE
|
||||||
| NC score | 0.030570 (rank : 149) | θ value | 1.06291 (rank : 100) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K1C9, Q91VX4 | Gene names | Lrrc41, Muf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 41 (Protein Muf1). | |||||
|
CJ082_HUMAN
|
||||||
| NC score | 0.029330 (rank : 150) | θ value | 2.36792 (rank : 110) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WW14 | Gene names | C10orf82 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf82. | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.028155 (rank : 151) | θ value | 1.38821 (rank : 103) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
TFAP4_HUMAN
|
||||||
| NC score | 0.027906 (rank : 152) | θ value | 0.365318 (rank : 85) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01664, O60409 | Gene names | TFAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-4 (Activating enhancer-binding protein 4). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.023654 (rank : 153) | θ value | 4.03905 (rank : 117) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.023108 (rank : 154) | θ value | 3.0926 (rank : 112) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
TRI42_HUMAN
|
||||||
| NC score | 0.021858 (rank : 155) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IWZ5, Q8N832, Q8NDL3 | Gene names | TRIM42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 42. | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.021726 (rank : 156) | θ value | 1.38821 (rank : 104) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
CXXC6_HUMAN
|
||||||
| NC score | 0.021270 (rank : 157) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFU7, Q5VUP7, Q7Z6B6, Q8TCR1, Q9C0I7 | Gene names | CXXC6, KIAA1676, LCX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CXXC-type zinc finger protein 6 (Leukemia-associated protein with a CXXC domain). | |||||
|
MEOX1_MOUSE
|
||||||
| NC score | 0.020832 (rank : 158) | θ value | 0.365318 (rank : 84) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P32442 | Gene names | Meox1, Mox-1, Mox1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-1 (Mesenchyme homeobox 1). | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.020767 (rank : 159) | θ value | 4.03905 (rank : 114) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
MEOX1_HUMAN
|
||||||
| NC score | 0.020124 (rank : 160) | θ value | 0.0736092 (rank : 66) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50221 | Gene names | MEOX1, MOX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein MOX-1 (Mesenchyme homeobox 1). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.020023 (rank : 161) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.019740 (rank : 162) | θ value | 4.03905 (rank : 116) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
XE7_HUMAN
|
||||||
| NC score | 0.018768 (rank : 163) | θ value | 0.365318 (rank : 87) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 738 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02040, Q02832 | Gene names | CXYorf3, DXYS155E, XE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-lymphocyte antigen precursor (B-lymphocyte surface antigen) (721P) (Protein XE7). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.016793 (rank : 164) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
GLE1_MOUSE
|
||||||
| NC score | 0.013813 (rank : 165) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8R322, Q3TT10, Q3TU23, Q3UD65, Q8BT16, Q9D4A6 | Gene names | Gle1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein). | |||||
|
CHM4A_HUMAN
|
||||||
| NC score | 0.013645 (rank : 166) | θ value | 2.36792 (rank : 109) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BY43, Q32Q79, Q86SZ8, Q96QJ9, Q9P026 | Gene names | CHMP4A, C14orf123, SHAX2 | |||
|
Domain Architecture |

|
|||||
| Description | Charged multivesicular body protein 4a (Chromatin-modifying protein 4a) (CHMP4a) (Vacuolar protein sorting 7-1) (SNF7-1) (hSnf-1) (SNF7 homolog associated with Alix-2). | |||||
|
SGOL2_MOUSE
|
||||||
| NC score | 0.012776 (rank : 167) | θ value | 5.27518 (rank : 124) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TSY8, Q811I4, Q8C9C4, Q8CGJ4, Q9CS55 | Gene names | Sgol2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Shugoshin-like 2. | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.012535 (rank : 168) | θ value | 1.06291 (rank : 93) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
PLCB3_MOUSE
|
||||||
| NC score | 0.012529 (rank : 169) | θ value | 1.38821 (rank : 107) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51432 | Gene names | Plcb3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.011492 (rank : 170) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
PLCB3_HUMAN
|
||||||
| NC score | 0.011088 (rank : 171) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q01970 | Gene names | PLCB3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 3 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-3) (PLC-beta-3). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.010689 (rank : 172) | θ value | 5.27518 (rank : 122) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SETBP_HUMAN
|
||||||
| NC score | 0.010664 (rank : 173) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y6X0, Q9UEF3 | Gene names | SETBP1, KIAA0437 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SET-binding protein (SEB). | |||||
|
URFB1_HUMAN
|
||||||
| NC score | 0.010021 (rank : 174) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6BDS2, Q9NXE0 | Gene names | UHRF1BP1, C6orf107 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UHRF1-binding protein 1 (Ubiquitin-like containing PHD and RING finger domains 1-binding protein 1) (ICBP90-binding protein 1). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.008546 (rank : 175) | θ value | 2.36792 (rank : 111) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.007362 (rank : 176) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.006408 (rank : 177) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.005712 (rank : 178) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.005435 (rank : 179) | θ value | 4.03905 (rank : 113) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
TBC12_HUMAN
|
||||||
| NC score | 0.004969 (rank : 180) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
POK9_HUMAN
|
||||||
| NC score | 0.004894 (rank : 181) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P63134 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_5q13.3 provirus ancestral Pol protein (HERV-K104 Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49)]. | |||||
|
UBP16_HUMAN
|
||||||
| NC score | 0.004727 (rank : 182) | θ value | 6.88961 (rank : 139) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5T5, Q8NEL3 | Gene names | USP16 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 16 (EC 3.1.2.15) (Ubiquitin thioesterase 16) (Ubiquitin-specific-processing protease 16) (Deubiquitinating enzyme 16) (Ubiquitin-processing protease UBP-M). | |||||
|
TSP2_MOUSE
|
||||||
| NC score | 0.003855 (rank : 183) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q03350 | Gene names | Thbs2, Tsp2 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-2 precursor. | |||||
|
UBE3A_HUMAN
|
||||||
| NC score | 0.002222 (rank : 184) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05086, P78355, Q93066, Q9UEP4, Q9UEP5, Q9UEP6, Q9UEP7, Q9UEP8, Q9UEP9 | Gene names | UBE3A, E6AP | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase E3A (EC 6.3.2.-) (E6AP ubiquitin-protein ligase) (Oncogenic protein-associated protein E6-AP) (Human papillomavirus E6-associated protein) (NY-REN-54 antigen). | |||||
|
POK12_HUMAN
|
||||||
| NC score | 0.002173 (rank : 185) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P63135 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Pol protein (HERV-K102 Pol protein) (HERV-K(III) Pol protein) [Includes: Reverse transcriptase (RT) (EC 2.7.7.49); Ribonuclease H (EC 3.1.26.4) (RNase H); Integrase (IN)]. | |||||
|
GCAB_MOUSE
|
||||||
| NC score | 0.001091 (rank : 186) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01864 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2A chain C region secreted form (B allele). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | -0.000126 (rank : 187) | θ value | 5.27518 (rank : 123) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | -0.001689 (rank : 188) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
LRRK2_HUMAN
|
||||||
| NC score | -0.002332 (rank : 189) | θ value | 1.06291 (rank : 101) | |||
| Query Neighborhood Hits | 148 | Target Neighborhood Hits | 1382 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5S007, Q6ZS50, Q8NCX9 | Gene names | LRRK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat serine/threonine-protein kinase 2 (EC 2.7.11.1) (Dardarin). | |||||