Please be patient as the page loads
|
TNR1B_HUMAN
|
||||||
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |
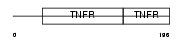
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TNR1B_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 3.87392e-144 (rank : 2) | NC score | 0.967517 (rank : 2) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
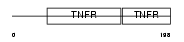
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
TNR6B_HUMAN
|
||||||
| θ value | 1.08979e-29 (rank : 3) | NC score | 0.880240 (rank : 3) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95407 | Gene names | TNFRSF6B, DCR3, TR6 | |||
|
Domain Architecture |
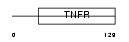
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6B precursor (Decoy receptor for Fas ligand) (Decoy receptor 3) (DcR3) (M68). | |||||
|
TR11B_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 4) | NC score | 0.863546 (rank : 4) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00300, O60236, Q53FX6, Q9UHP4 | Gene names | TNFRSF11B, OCIF, OPG | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TR11B_MOUSE
|
||||||
| θ value | 2.51237e-26 (rank : 5) | NC score | 0.853200 (rank : 6) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08712, O70202 | Gene names | Tnfrsf11b, Ocif, Opg | |||
|
Domain Architecture |
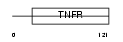
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR3_HUMAN
|
||||||
| θ value | 3.28125e-26 (rank : 6) | NC score | 0.855134 (rank : 5) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
TNR21_HUMAN
|
||||||
| θ value | 1.24688e-25 (rank : 7) | NC score | 0.792048 (rank : 13) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR5_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 8) | NC score | 0.843920 (rank : 7) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P25942, Q9BYU0 | Gene names | CD40, TNFRSF5 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (CDw40) (Bp50). | |||||
|
TNR21_MOUSE
|
||||||
| θ value | 8.08199e-25 (rank : 9) | NC score | 0.784331 (rank : 14) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR3_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 10) | NC score | 0.834768 (rank : 9) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P50284 | Gene names | Ltbr, Tnfcr, Tnfrsf3 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor). | |||||
|
TNR11_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 11) | NC score | 0.796432 (rank : 12) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
TNR11_MOUSE
|
||||||
| θ value | 9.24701e-21 (rank : 12) | NC score | 0.798148 (rank : 11) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
TNR14_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 13) | NC score | 0.836157 (rank : 8) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
TNR5_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 14) | NC score | 0.822278 (rank : 10) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
TNR8_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 15) | NC score | 0.683610 (rank : 23) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TNR4_HUMAN
|
||||||
| θ value | 1.58096e-12 (rank : 16) | NC score | 0.741717 (rank : 16) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
TNR4_MOUSE
|
||||||
| θ value | 3.52202e-12 (rank : 17) | NC score | 0.734042 (rank : 17) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P47741 | Gene names | Tnfrsf4, Ox40, Txgp1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (OX40 antigen). | |||||
|
TNR16_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 18) | NC score | 0.678477 (rank : 24) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR22_MOUSE
|
||||||
| θ value | 3.89403e-11 (rank : 19) | NC score | 0.730961 (rank : 18) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ER62, Q8VHB9, Q9CZA4 | Gene names | Tnfrsf22, Dctrailr2, Tnfrh2, Tnfrsf1al2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 22 (Tumor necrosis factor receptor p60 homolog 2) (TNF receptor family member SOBa) (Decoy TRAIL receptor 2) (TNF receptor homolog 2). | |||||
|
TNR16_MOUSE
|
||||||
| θ value | 1.47974e-10 (rank : 20) | NC score | 0.684591 (rank : 22) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
TNR1A_MOUSE
|
||||||
| θ value | 1.9326e-10 (rank : 21) | NC score | 0.667973 (rank : 25) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR9_MOUSE
|
||||||
| θ value | 3.29651e-10 (rank : 22) | NC score | 0.700873 (rank : 20) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
TNR9_HUMAN
|
||||||
| θ value | 1.25267e-09 (rank : 23) | NC score | 0.688560 (rank : 21) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
TNR26_MOUSE
|
||||||
| θ value | 1.63604e-09 (rank : 24) | NC score | 0.748947 (rank : 15) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P83626 | Gene names | Tnfrsf26, Tnfrh3 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 26 precursor (TNF receptor homolog 3). | |||||
|
TNR1A_HUMAN
|
||||||
| θ value | 1.80886e-08 (rank : 25) | NC score | 0.646508 (rank : 26) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
TNR23_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 26) | NC score | 0.715485 (rank : 19) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9ER63, Q8VHC0 | Gene names | Tnfrsf23, Dctrailr1, Tnfrh1, Tnfrsf1al1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 23 precursor (Tumor necrosis factor receptor p60 homolog 1) (TNF receptor family member SOB) (Decoy TRAIL receptor 1) (TNF receptor homolog 1). | |||||
|
TNR6_MOUSE
|
||||||
| θ value | 6.87365e-08 (rank : 27) | NC score | 0.646265 (rank : 27) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P25446, Q9DCQ1 | Gene names | Fas, Apt1, Tnfrsf6 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TR10A_HUMAN
|
||||||
| θ value | 1.53129e-07 (rank : 28) | NC score | 0.500846 (rank : 35) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
TNR6_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 29) | NC score | 0.613362 (rank : 28) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P25445, Q14292, Q14293, Q14294, Q14295, Q16652, Q6SSE9 | Gene names | FAS, APT1, FAS1, TNFRSF6 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR8_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 30) | NC score | 0.553549 (rank : 31) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
TR10B_HUMAN
|
||||||
| θ value | 4.45536e-07 (rank : 31) | NC score | 0.530298 (rank : 32) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
TNR7_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 32) | NC score | 0.573067 (rank : 29) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
TNR25_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 33) | NC score | 0.562001 (rank : 30) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 34) | NC score | 0.524541 (rank : 34) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
TR10D_HUMAN
|
||||||
| θ value | 1.87187e-05 (rank : 35) | NC score | 0.487613 (rank : 37) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBN6, Q9Y6Q4 | Gene names | TNFRSF10D, DCR2, TRAILR4, TRUNDD | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10D precursor (Decoy receptor 2) (DcR2) (TNF-related apoptosis-inducing ligand receptor 4) (TRAIL receptor 4) (TRAIL-R4) (TRAIL receptor with a truncated death domain) (CD264 antigen). | |||||
|
TNR27_HUMAN
|
||||||
| θ value | 3.19293e-05 (rank : 36) | NC score | 0.391896 (rank : 39) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HAV5, Q6UWM2, Q8IZA6 | Gene names | EDA2R, TNFRSF27, XEDAR | |||
|
Domain Architecture |
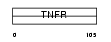
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TNR18_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 37) | NC score | 0.489123 (rank : 36) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
|
TNR27_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 38) | NC score | 0.340032 (rank : 41) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BX35, Q8BM50 | Gene names | Eda2r, Tnfrsf27, Xedar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TNR7_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 39) | NC score | 0.527101 (rank : 33) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
TR10B_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 40) | NC score | 0.464201 (rank : 38) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QZM4, Q9JJL5, Q9JJL6 | Gene names | Tnfrsf10b, Dr5, Killer | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (MK). | |||||
|
LAMC3_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 41) | NC score | 0.140298 (rank : 49) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
SREC2_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 42) | NC score | 0.103666 (rank : 62) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 43) | NC score | 0.130233 (rank : 51) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 44) | NC score | 0.157803 (rank : 48) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 45) | NC score | 0.119073 (rank : 54) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
EDAR_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 46) | NC score | 0.205014 (rank : 47) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
FGRL1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 47) | NC score | 0.027868 (rank : 159) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V87, Q920C2, Q920C3 | Gene names | Fgfrl1, Fgfr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5). | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 48) | NC score | 0.109594 (rank : 57) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
PCSK6_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 49) | NC score | 0.108643 (rank : 59) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 50) | NC score | 0.067591 (rank : 104) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
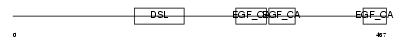
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 51) | NC score | 0.111041 (rank : 55) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
TR19L_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 52) | NC score | 0.298941 (rank : 43) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |
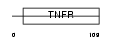
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
FURIN_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 53) | NC score | 0.063628 (rank : 112) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23188 | Gene names | Furin, Fur, Pcsk3 | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme) (Prohormone convertase 3). | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 54) | NC score | 0.054528 (rank : 132) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NTNG2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 55) | NC score | 0.081838 (rank : 82) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R4F1, Q8R4F2, Q8VIP6, Q8VIP7, Q8VIP8 | Gene names | Ntng2, Lmnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
TEX14_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 56) | NC score | 0.010882 (rank : 181) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
TNR18_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 57) | NC score | 0.375368 (rank : 40) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35714, Q9JKR1, Q9JKR2, Q9JKR3 | Gene names | Tnfrsf18, Gitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein). | |||||
|
EDAR_HUMAN
|
||||||
| θ value | 0.125558 (rank : 58) | NC score | 0.207235 (rank : 46) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
FGRL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 59) | NC score | 0.024899 (rank : 162) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N441, Q6PJN1, Q9BXN7, Q9H4D7 | Gene names | FGFRL1, FGFR5, FHFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5) (FGFR-like protein) (FGF homologous factor receptor). | |||||
|
TR19L_MOUSE
|
||||||
| θ value | 0.125558 (rank : 60) | NC score | 0.299658 (rank : 42) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BX43, Q8BTV0 | Gene names | Tnfrsf19l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor. | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | 0.163984 (rank : 61) | NC score | 0.126880 (rank : 52) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 62) | NC score | 0.130835 (rank : 50) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 63) | NC score | 0.016052 (rank : 170) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 64) | NC score | 0.124582 (rank : 53) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 65) | NC score | 0.096096 (rank : 70) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | 0.279714 (rank : 66) | NC score | 0.054834 (rank : 131) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
FURIN_HUMAN
|
||||||
| θ value | 0.365318 (rank : 67) | NC score | 0.064400 (rank : 109) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09958, Q14336, Q6LBS3 | Gene names | FURIN, FUR, PACE | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme). | |||||
|
GPC6A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 68) | NC score | 0.014529 (rank : 176) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
SREC_HUMAN
|
||||||
| θ value | 0.62314 (rank : 69) | NC score | 0.107430 (rank : 60) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.813845 (rank : 70) | NC score | 0.022064 (rank : 163) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DLL4_MOUSE
|
||||||
| θ value | 0.813845 (rank : 71) | NC score | 0.061956 (rank : 121) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
ESM1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 72) | NC score | 0.063069 (rank : 115) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
TIF1B_HUMAN
|
||||||
| θ value | 0.813845 (rank : 73) | NC score | 0.026180 (rank : 160) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
AIRE_MOUSE
|
||||||
| θ value | 1.06291 (rank : 74) | NC score | 0.021673 (rank : 165) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
SIVA_MOUSE
|
||||||
| θ value | 1.06291 (rank : 75) | NC score | 0.035456 (rank : 153) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54926, Q3U6J2, Q921I8, Q9CWL1, Q9R1Q0, Q9R1Q1 | Gene names | Siva | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 76) | NC score | 0.096350 (rank : 69) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
TIF1B_MOUSE
|
||||||
| θ value | 1.06291 (rank : 77) | NC score | 0.024969 (rank : 161) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
USH2A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 78) | NC score | 0.094985 (rank : 71) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
TNR19_HUMAN
|
||||||
| θ value | 1.38821 (rank : 79) | NC score | 0.234838 (rank : 45) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NS68, Q9BXZ9, Q9BY00, Q9NZV2 | Gene names | TNFRSF19, TAJ, TROY | |||
|
Domain Architecture |
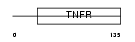
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
JARD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 80) | NC score | 0.015860 (rank : 171) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 1.81305 (rank : 81) | NC score | 0.084097 (rank : 80) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.021814 (rank : 164) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EPHB4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.006875 (rank : 186) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54760, Q9BTA5, Q9BXP0 | Gene names | EPHB4, HTK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HTK). | |||||
|
FAM5B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 84) | NC score | 0.014888 (rank : 175) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9C0B6, O95560, Q6ZWC1, Q7LCZ9, Q8N360 | Gene names | FAM5B, BRINP2, DBCCR1L2, KIAA1747 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5B precursor (BMP/retinoic acid-inducible neural-specific protein 2) (DBCCR1-like protein 2). | |||||
|
KR109_HUMAN
|
||||||
| θ value | 2.36792 (rank : 85) | NC score | 0.052999 (rank : 134) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
STON1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 86) | NC score | 0.015736 (rank : 172) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
TNR6A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 87) | NC score | 0.031177 (rank : 157) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
ZPBP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 88) | NC score | 0.040612 (rank : 151) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6X786, Q6X785, Q9DA91 | Gene names | Zpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida-binding protein 2 precursor. | |||||
|
FHL2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.015057 (rank : 174) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70433, P97448 | Gene names | Fhl2 | |||
|
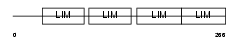
Domain Architecture |
|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.012834 (rank : 178) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
KRA59_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.093880 (rank : 74) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.099822 (rank : 65) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.047516 (rank : 148) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.066184 (rank : 108) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SLIT3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 95) | NC score | 0.031546 (rank : 156) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
CBL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.008714 (rank : 183) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.062566 (rank : 118) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.064251 (rank : 111) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.093098 (rank : 75) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.019872 (rank : 167) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
PER3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.010009 (rank : 182) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.007828 (rank : 184) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TSP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.032176 (rank : 154) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
VINEX_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.006402 (rank : 188) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
FHL3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 105) | NC score | 0.018053 (rank : 168) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13643 | Gene names | FHL3, SLIM2 | |||
|
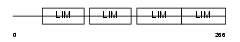
Domain Architecture |
|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.069613 (rank : 99) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.099978 (rank : 64) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.011548 (rank : 180) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
TSP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.031971 (rank : 155) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 110) | NC score | 0.013691 (rank : 177) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
DLL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 111) | NC score | 0.058802 (rank : 124) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 112) | NC score | 0.042645 (rank : 150) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 6.88961 (rank : 113) | NC score | 0.005231 (rank : 189) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 6.88961 (rank : 114) | NC score | 0.012818 (rank : 179) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 115) | NC score | 0.052538 (rank : 135) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
FHL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.020528 (rank : 166) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R059, Q9JLP5, Q9WUH3 | Gene names | Fhl3 | |||
|
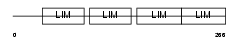
Domain Architecture |
|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
GP179_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.017539 (rank : 169) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.074085 (rank : 90) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 119) | NC score | 0.109724 (rank : 56) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
LRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 120) | NC score | 0.037463 (rank : 152) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 121) | NC score | 0.047417 (rank : 149) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 122) | NC score | 0.006827 (rank : 187) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
NELL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 123) | NC score | 0.048852 (rank : 147) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
SLIT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 124) | NC score | 0.029816 (rank : 158) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
TCF20_MOUSE
|
||||||
| θ value | 8.99809 (rank : 125) | NC score | 0.007545 (rank : 185) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 8.99809 (rank : 126) | NC score | 0.015148 (rank : 173) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
USH2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 127) | NC score | 0.087737 (rank : 79) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.063270 (rank : 114) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ATRN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.050837 (rank : 142) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
ATRN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052065 (rank : 136) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
CREL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.059219 (rank : 123) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.054980 (rank : 129) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
DLK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.050956 (rank : 141) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
DLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.057924 (rank : 126) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.098039 (rank : 68) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.050505 (rank : 145) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
KR101_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.062286 (rank : 119) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
KR102_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051032 (rank : 140) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
KR106_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.057756 (rank : 127) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR510_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.057421 (rank : 128) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
KR511_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.050123 (rank : 146) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.075902 (rank : 88) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.069637 (rank : 98) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.078050 (rank : 85) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.063606 (rank : 113) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.066330 (rank : 107) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.054863 (rank : 130) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.067633 (rank : 103) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.099482 (rank : 66) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.091854 (rank : 76) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.082678 (rank : 81) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.088655 (rank : 77) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.072289 (rank : 93) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.076365 (rank : 87) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.088404 (rank : 78) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.094659 (rank : 72) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.098147 (rank : 67) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.079307 (rank : 84) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
LAMB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.076655 (rank : 86) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAMC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.103573 (rank : 63) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.079799 (rank : 83) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.107177 (rank : 61) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.062587 (rank : 117) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28184 | Gene names | Mt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF). | |||||
|
NET1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.062149 (rank : 120) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95631 | Gene names | NTN1, NTN1L | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
NET1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.062930 (rank : 116) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09118, Q60832, Q9QY50 | Gene names | Ntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
NET2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.068083 (rank : 102) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
NET2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.067169 (rank : 105) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9R1A3, Q9QY49, Q9WVA6 | Gene names | Ntn2l, Ntn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor (Netrin-3). | |||||
|
NET4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.071166 (rank : 97) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
NET4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.067000 (rank : 106) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JI33 | Gene names | Ntn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin). | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.050616 (rank : 144) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NTNG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.071406 (rank : 95) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.072100 (rank : 94) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.071220 (rank : 96) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CW9, Q6UXY0, Q96JH0 | Gene names | NTNG2, KIAA1857, LMNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
PCSK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.050619 (rank : 143) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UW60, Q8IY88, Q9UF79 | Gene names | PCSK4, PC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (Proprotein convertase PC4). | |||||
|
PCSK5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.108682 (rank : 58) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.073204 (rank : 91) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
RSPO1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.058675 (rank : 125) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q2MKA7, Q5T0F2, Q8N7L5 | Gene names | RSPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (hRspo1). | |||||
|
RSPO4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053386 (rank : 133) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.072872 (rank : 92) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.074473 (rank : 89) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.051891 (rank : 138) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.068975 (rank : 100) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.068109 (rank : 101) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.061829 (rank : 122) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.064358 (rank : 110) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TNR19_MOUSE
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.294952 (rank : 44) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLL3, Q812G3, Q9JHF1, Q9JJH6, Q9JLL2, Q9QXW7 | Gene names | Tnfrsf19, Taj, Troy | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.051479 (rank : 139) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.051923 (rank : 137) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.093921 (rank : 73) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TNR1B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 127 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.967517 (rank : 2) | θ value | 3.87392e-144 (rank : 2) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
TNR6B_HUMAN
|
||||||
| NC score | 0.880240 (rank : 3) | θ value | 1.08979e-29 (rank : 3) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O95407 | Gene names | TNFRSF6B, DCR3, TR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6B precursor (Decoy receptor for Fas ligand) (Decoy receptor 3) (DcR3) (M68). | |||||
|
TR11B_HUMAN
|
||||||
| NC score | 0.863546 (rank : 4) | θ value | 1.85889e-29 (rank : 4) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O00300, O60236, Q53FX6, Q9UHP4 | Gene names | TNFRSF11B, OCIF, OPG | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR3_HUMAN
|
||||||
| NC score | 0.855134 (rank : 5) | θ value | 3.28125e-26 (rank : 6) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
TR11B_MOUSE
|
||||||
| NC score | 0.853200 (rank : 6) | θ value | 2.51237e-26 (rank : 5) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O08712, O70202 | Gene names | Tnfrsf11b, Ocif, Opg | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR5_HUMAN
|
||||||
| NC score | 0.843920 (rank : 7) | θ value | 2.12685e-25 (rank : 8) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P25942, Q9BYU0 | Gene names | CD40, TNFRSF5 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (CDw40) (Bp50). | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.836157 (rank : 8) | θ value | 1.5773e-20 (rank : 13) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
TNR3_MOUSE
|
||||||
| NC score | 0.834768 (rank : 9) | θ value | 7.56453e-23 (rank : 10) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P50284 | Gene names | Ltbr, Tnfcr, Tnfrsf3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor). | |||||
|
TNR5_MOUSE
|
||||||
| NC score | 0.822278 (rank : 10) | θ value | 1.74391e-19 (rank : 14) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
TNR11_MOUSE
|
||||||
| NC score | 0.798148 (rank : 11) | θ value | 9.24701e-21 (rank : 12) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
TNR11_HUMAN
|
||||||
| NC score | 0.796432 (rank : 12) | θ value | 9.87957e-23 (rank : 11) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
TNR21_HUMAN
|
||||||
| NC score | 0.792048 (rank : 13) | θ value | 1.24688e-25 (rank : 7) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR21_MOUSE
|
||||||
| NC score | 0.784331 (rank : 14) | θ value | 8.08199e-25 (rank : 9) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR26_MOUSE
|
||||||
| NC score | 0.748947 (rank : 15) | θ value | 1.63604e-09 (rank : 24) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P83626 | Gene names | Tnfrsf26, Tnfrh3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 26 precursor (TNF receptor homolog 3). | |||||
|
TNR4_HUMAN
|
||||||
| NC score | 0.741717 (rank : 16) | θ value | 1.58096e-12 (rank : 16) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
TNR4_MOUSE
|
||||||
| NC score | 0.734042 (rank : 17) | θ value | 3.52202e-12 (rank : 17) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P47741 | Gene names | Tnfrsf4, Ox40, Txgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (OX40 antigen). | |||||
|
TNR22_MOUSE
|
||||||
| NC score | 0.730961 (rank : 18) | θ value | 3.89403e-11 (rank : 19) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9ER62, Q8VHB9, Q9CZA4 | Gene names | Tnfrsf22, Dctrailr2, Tnfrh2, Tnfrsf1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 22 (Tumor necrosis factor receptor p60 homolog 2) (TNF receptor family member SOBa) (Decoy TRAIL receptor 2) (TNF receptor homolog 2). | |||||
|
TNR23_MOUSE
|
||||||
| NC score | 0.715485 (rank : 19) | θ value | 6.87365e-08 (rank : 26) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9ER63, Q8VHC0 | Gene names | Tnfrsf23, Dctrailr1, Tnfrh1, Tnfrsf1al1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 23 precursor (Tumor necrosis factor receptor p60 homolog 1) (TNF receptor family member SOB) (Decoy TRAIL receptor 1) (TNF receptor homolog 1). | |||||
|
TNR9_MOUSE
|
||||||
| NC score | 0.700873 (rank : 20) | θ value | 3.29651e-10 (rank : 22) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
TNR9_HUMAN
|
||||||
| NC score | 0.688560 (rank : 21) | θ value | 1.25267e-09 (rank : 23) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
TNR16_MOUSE
|
||||||
| NC score | 0.684591 (rank : 22) | θ value | 1.47974e-10 (rank : 20) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.683610 (rank : 23) | θ value | 1.99529e-15 (rank : 15) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.678477 (rank : 24) | θ value | 7.84624e-12 (rank : 18) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR1A_MOUSE
|
||||||
| NC score | 0.667973 (rank : 25) | θ value | 1.9326e-10 (rank : 21) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR1A_HUMAN
|
||||||
| NC score | 0.646508 (rank : 26) | θ value | 1.80886e-08 (rank : 25) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
TNR6_MOUSE
|
||||||
| NC score | 0.646265 (rank : 27) | θ value | 6.87365e-08 (rank : 27) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | P25446, Q9DCQ1 | Gene names | Fas, Apt1, Tnfrsf6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR6_HUMAN
|
||||||
| NC score | 0.613362 (rank : 28) | θ value | 1.99992e-07 (rank : 29) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P25445, Q14292, Q14293, Q14294, Q14295, Q16652, Q6SSE9 | Gene names | FAS, APT1, FAS1, TNFRSF6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR7_HUMAN
|
||||||
| NC score | 0.573067 (rank : 29) | θ value | 8.40245e-06 (rank : 32) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.562001 (rank : 30) | θ value | 1.43324e-05 (rank : 33) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.553549 (rank : 31) | θ value | 2.61198e-07 (rank : 30) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
TR10B_HUMAN
|
||||||
| NC score | 0.530298 (rank : 32) | θ value | 4.45536e-07 (rank : 31) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
TNR7_MOUSE
|
||||||
| NC score | 0.527101 (rank : 33) | θ value | 0.00175202 (rank : 39) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.524541 (rank : 34) | θ value | 1.87187e-05 (rank : 34) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
TR10A_HUMAN
|
||||||
| NC score | 0.500846 (rank : 35) | θ value | 1.53129e-07 (rank : 28) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
TNR18_HUMAN
|
||||||
| NC score | 0.489123 (rank : 36) | θ value | 0.000270298 (rank : 37) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
|
TR10D_HUMAN
|
||||||
| NC score | 0.487613 (rank : 37) | θ value | 1.87187e-05 (rank : 35) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9UBN6, Q9Y6Q4 | Gene names | TNFRSF10D, DCR2, TRAILR4, TRUNDD | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10D precursor (Decoy receptor 2) (DcR2) (TNF-related apoptosis-inducing ligand receptor 4) (TRAIL receptor 4) (TRAIL-R4) (TRAIL receptor with a truncated death domain) (CD264 antigen). | |||||
|
TR10B_MOUSE
|
||||||
| NC score | 0.464201 (rank : 38) | θ value | 0.00390308 (rank : 40) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9QZM4, Q9JJL5, Q9JJL6 | Gene names | Tnfrsf10b, Dr5, Killer | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (MK). | |||||
|
TNR27_HUMAN
|
||||||
| NC score | 0.391896 (rank : 39) | θ value | 3.19293e-05 (rank : 36) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HAV5, Q6UWM2, Q8IZA6 | Gene names | EDA2R, TNFRSF27, XEDAR | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TNR18_MOUSE
|
||||||
| NC score | 0.375368 (rank : 40) | θ value | 0.0961366 (rank : 57) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35714, Q9JKR1, Q9JKR2, Q9JKR3 | Gene names | Tnfrsf18, Gitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein). | |||||
|
TNR27_MOUSE
|
||||||
| NC score | 0.340032 (rank : 41) | θ value | 0.000786445 (rank : 38) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BX35, Q8BM50 | Gene names | Eda2r, Tnfrsf27, Xedar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TR19L_MOUSE
|
||||||
| NC score | 0.299658 (rank : 42) | θ value | 0.125558 (rank : 60) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8BX43, Q8BTV0 | Gene names | Tnfrsf19l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor. | |||||
|
TR19L_HUMAN
|
||||||
| NC score | 0.298941 (rank : 43) | θ value | 0.0736092 (rank : 52) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
TNR19_MOUSE
|
||||||
| NC score | 0.294952 (rank : 44) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9JLL3, Q812G3, Q9JHF1, Q9JJH6, Q9JLL2, Q9QXW7 | Gene names | Tnfrsf19, Taj, Troy | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
TNR19_HUMAN
|
||||||
| NC score | 0.234838 (rank : 45) | θ value | 1.38821 (rank : 79) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NS68, Q9BXZ9, Q9BY00, Q9NZV2 | Gene names | TNFRSF19, TAJ, TROY | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
EDAR_HUMAN
|
||||||
| NC score | 0.207235 (rank : 46) | θ value | 0.125558 (rank : 58) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
EDAR_MOUSE
|
||||||
| NC score | 0.205014 (rank : 47) | θ value | 0.0252991 (rank : 46) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.157803 (rank : 48) | θ value | 0.0148317 (rank : 44) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 92 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
LAMC3_MOUSE
|
||||||
| NC score | 0.140298 (rank : 49) | θ value | 0.0113563 (rank : 41) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.130835 (rank : 50) | θ value | 0.163984 (rank : 62) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.130233 (rank : 51) | θ value | 0.0148317 (rank : 43) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.126880 (rank : 52) | θ value | 0.163984 (rank : 61) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.124582 (rank : 53) | θ value | 0.21417 (rank : 64) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 80 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.119073 (rank : 54) | θ value | 0.0193708 (rank : 45) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.111041 (rank : 55) | θ value | 0.0736092 (rank : 51) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.109724 (rank : 56) | θ value | 8.99809 (rank : 119) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.109594 (rank : 57) | θ value | 0.0330416 (rank : 48) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
PCSK5_HUMAN
|
||||||
| NC score | 0.108682 (rank : 58) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q92824, Q13527 | Gene names | PCSK5, PC5, PC6 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (hPC6). | |||||
|
PCSK6_HUMAN
|
||||||
| NC score | 0.108643 (rank : 59) | θ value | 0.0431538 (rank : 49) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P29122, Q15099, Q15100, Q9UEG7, Q9UEJ1, Q9UEJ2, Q9UEJ7, Q9UEJ8, Q9UEJ9, Q9Y4G9, Q9Y4H0, Q9Y4H1 | Gene names | PCSK6, PACE4 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 6 precursor (EC 3.4.21.-) (Paired basic amino acid cleaving enzyme 4) (Subtilisin/kexin-like protease PACE4) (Subtilisin-like proprotein convertase 4) (SPC4). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.107430 (rank : 60) | θ value | 0.62314 (rank : 69) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.107177 (rank : 61) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.103666 (rank : 62) | θ value | 0.0113563 (rank : 42) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
LAMC1_MOUSE
|
||||||
| NC score | 0.103573 (rank : 63) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.099978 (rank : 64) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.099822 (rank : 65) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.099482 (rank : 66) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.098147 (rank : 67) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.098039 (rank : 68) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.096350 (rank : 69) | θ value | 1.06291 (rank : 76) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.096096 (rank : 70) | θ value | 0.279714 (rank : 65) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.094985 (rank : 71) | θ value | 1.06291 (rank : 78) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
LAMB2_HUMAN
|
||||||
| NC score | 0.094659 (rank : 72) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.093921 (rank : 73) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.093880 (rank : 74) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.093098 (rank : 75) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.091854 (rank : 76) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.088655 (rank : 77) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.088404 (rank : 78) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.087737 (rank : 79) | θ value | 8.99809 (rank : 127) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.084097 (rank : 80) | θ value | 1.81305 (rank : 81) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.082678 (rank : 81) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
NTNG2_MOUSE
|
||||||
| NC score | 0.081838 (rank : 82) | θ value | 0.0961366 (rank : 55) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R4F1, Q8R4F2, Q8VIP6, Q8VIP7, Q8VIP8 | Gene names | Ntng2, Lmnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.079799 (rank : 83) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.079307 (rank : 84) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.078050 (rank : 85) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
LAMB3_MOUSE
|
||||||
| NC score | 0.076655 (rank : 86) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAMA4_MOUSE
|
||||||
| NC score | 0.076365 (rank : 87) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.075902 (rank : 88) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.074473 (rank : 89) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.074085 (rank : 90) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.073204 (rank : 91) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.072872 (rank : 92) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.072289 (rank : 93) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
NTNG1_MOUSE
|
||||||
| NC score | 0.072100 (rank : 94) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_HUMAN
|
||||||
| NC score | 0.071406 (rank : 95) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG2_HUMAN
|
||||||
| NC score | 0.071220 (rank : 96) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96CW9, Q6UXY0, Q96JH0 | Gene names | NTNG2, KIAA1857, LMNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
NET4_HUMAN
|
||||||
| NC score | 0.071166 (rank : 97) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
KRA52_HUMAN
|
||||||
| NC score | 0.069637 (rank : 98) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q701N4 | Gene names | KRTAP5-2, KAP5-8, KAP5.2, KRTAP5-8, KRTAP5.2, KRTAP5.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-2 (Keratin-associated protein 5.2) (Ultrahigh sulfur keratin-associated protein 5.2) (Keratin-associated protein 5-8) (Keratin-associated protein 5.8). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.069613 (rank : 99) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.068975 (rank : 100) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.068109 (rank : 101) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
NET2_HUMAN
|
||||||
| NC score | 0.068083 (rank : 102) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O00634 | Gene names | NTN2L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor. | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.067633 (rank : 103) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.067591 (rank : 104) | θ value | 0.0736092 (rank : 50) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
NET2_MOUSE
|
||||||
| NC score | 0.067169 (rank : 105) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9R1A3, Q9QY49, Q9WVA6 | Gene names | Ntn2l, Ntn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-2-like protein precursor (Netrin-3). | |||||
|
NET4_MOUSE
|
||||||
| NC score | 0.067000 (rank : 106) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9JI33 | Gene names | Ntn4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.066330 (rank : 107) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.066184 (rank : 108) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
FURIN_HUMAN
|
||||||
| NC score | 0.064400 (rank : 109) | θ value | 0.365318 (rank : 67) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09958, Q14336, Q6LBS3 | Gene names | FURIN, FUR, PACE | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme). | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.064358 (rank : 110) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.064251 (rank : 111) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
FURIN_MOUSE
|
||||||
| NC score | 0.063628 (rank : 112) | θ value | 0.0961366 (rank : 53) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23188 | Gene names | Furin, Fur, Pcsk3 | |||
|
Domain Architecture |

|
|||||
| Description | Furin precursor (EC 3.4.21.75) (Paired basic amino acid residue cleaving enzyme) (PACE) (Dibasic-processing enzyme) (Prohormone convertase 3). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.063606 (rank : 113) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.063270 (rank : 114) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
ESM1_HUMAN
|
||||||
| NC score | 0.063069 (rank : 115) | θ value | 0.813845 (rank : 72) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NQ30, Q15330, Q96ES3 | Gene names | ESM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cell-specific molecule 1 precursor (ESM-1 secretory protein) (ESM-1). | |||||
|
NET1_MOUSE
|
||||||
| NC score | 0.062930 (rank : 116) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O09118, Q60832, Q9QY50 | Gene names | Ntn1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
MT3_MOUSE
|
||||||
| NC score | 0.062587 (rank : 117) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P28184 | Gene names | Mt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF). | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.062566 (rank : 118) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
KR101_HUMAN
|
||||||
| NC score | 0.062286 (rank : 119) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P60331 | Gene names | KRTAP10-1, KAP10.1, KAP18-1, KRTAP10.1, KRTAP18-1, KRTAP18.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-1 (Keratin-associated protein 10.1) (High sulfur keratin-associated protein 10.1) (Keratin-associated protein 18-1) (Keratin-associated protein 18.1). | |||||
|
NET1_HUMAN
|
||||||
| NC score | 0.062149 (rank : 120) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O95631 | Gene names | NTN1, NTN1L | |||
|
Domain Architecture |

|
|||||
| Description | Netrin-1 precursor. | |||||
|
DLL4_MOUSE
|
||||||
| NC score | 0.061956 (rank : 121) | θ value | 0.813845 (rank : 71) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9JI71, Q9JHZ7 | Gene names | Dll4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.061829 (rank : 122) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.059219 (rank : 123) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.058802 (rank : 124) | θ value | 6.88961 (rank : 111) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
RSPO1_HUMAN
|
||||||
| NC score | 0.058675 (rank : 125) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q2MKA7, Q5T0F2, Q8N7L5 | Gene names | RSPO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-1 precursor (Roof plate-specific spondin-1) (hRspo1). | |||||
|
DLL1_HUMAN
|
||||||
| NC score | 0.057924 (rank : 126) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O00548, Q9NU41, Q9UJV2 | Gene names | DLL1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1) (H-Delta-1). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.057756 (rank : 127) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR510_HUMAN
|
||||||
| NC score | 0.057421 (rank : 128) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8G5 | Gene names | KRTAP5-10, KAP5.10, KRTAP5.10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-10 (Keratin-associated protein 5.10) (Ultrahigh sulfur keratin-associated protein 5.10). | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.054980 (rank : 129) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.054863 (rank : 130) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.054834 (rank : 131) | θ value | 0.279714 (rank : 66) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.054528 (rank : 132) | θ value | 0.0961366 (rank : 54) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
RSPO4_HUMAN
|
||||||
| NC score | 0.053386 (rank : 133) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q2I0M5, Q9UGB2 | Gene names | RSPO4, C20orf182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R-spondin-4 precursor (Roof plate-specific spondin-4) (hRspo4). | |||||
|
KR109_HUMAN
|
||||||
| NC score | 0.052999 (rank : 134) | θ value | 2.36792 (rank : 85) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P60411, Q70LJ1 | Gene names | KRTAP10-9, KAP10.9, KAP18-9, KRTAP10.9, KRTAP18-9, KRTAP18.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-9 (Keratin-associated protein 10.9) (High sulfur keratin-associated protein 10.9) (Keratin-associated protein 18-9) (Keratin-associated protein 18.9). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.052538 (rank : 135) | θ value | 6.88961 (rank : 115) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
ATRN_MOUSE
|
||||||
| NC score | 0.052065 (rank : 136) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9WU60, Q9R263, Q9WU77 | Gene names | Atrn, mg, Mgca | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany protein). | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.051923 (rank : 137) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.051891 (rank : 138) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.051479 (rank : 139) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
KR102_HUMAN
|
||||||
| NC score | 0.051032 (rank : 140) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P60368, Q70LJ5 | Gene names | KRTAP10-2, KAP10.2, KAP18-2, KRTAP10.2, KRTAP18-2, KRTAP18.2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-2 (Keratin-associated protein 10.2) (High sulfur keratin-associated protein 10.2) (Keratin-associated protein 18-2) (Keratin-associated protein 18.2). | |||||
|
DLK_HUMAN
|
||||||
| NC score | 0.050956 (rank : 141) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P80370, P15803, Q96DW5 | Gene names | DLK1, DLK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Delta-like protein precursor (DLK) (pG2) [Contains: Fetal antigen 1 (FA1)]. | |||||
|
ATRN_HUMAN
|
||||||
| NC score | 0.050837 (rank : 142) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 525 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O75882, O60295, O95414, Q3MIT3, Q5TDA2, Q5TDA4, Q5VYW3, Q9NTQ3, Q9NTQ4, Q9NU01, Q9NZ57, Q9NZ58 | Gene names | ATRN, KIAA0548, MGCA | |||
|
Domain Architecture |

|
|||||
| Description | Attractin precursor (Mahogany homolog) (DPPT-L). | |||||
|
PCSK4_HUMAN
|
||||||
| NC score | 0.050619 (rank : 143) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6UW60, Q8IY88, Q9UF79 | Gene names | PCSK4, PC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proprotein convertase subtilisin/kexin type 4 precursor (EC 3.4.21.-) (Proprotein convertase PC4). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.050616 (rank : 144) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.050505 (rank : 145) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
KR511_HUMAN
|
||||||
| NC score | 0.050123 (rank : 146) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6L8G4 | Gene names | KRTAP5-11, KAP5.11, KRTAP5.11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-11 (Keratin-associated protein 5.11) (Ultrahigh sulfur keratin-associated protein 5.11). | |||||
|
NELL1_HUMAN
|
||||||
| NC score | 0.048852 (rank : 147) | θ value | 8.99809 (rank : 123) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q92832, Q9Y472 | Gene names | NELL1, NRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL1 precursor (NEL-like protein 1) (Nel-related protein 1). | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.047516 (rank : 148) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.047417 (rank : 149) | θ value | 8.99809 (rank : 121) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.042645 (rank : 150) | θ value | 6.88961 (rank : 112) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
ZPBP2_MOUSE
|
||||||
| NC score | 0.040612 (rank : 151) | θ value | 2.36792 (rank : 88) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6X786, Q6X785, Q9DA91 | Gene names | Zpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zona pellucida-binding protein 2 precursor. | |||||
|
LRP1_HUMAN
|
||||||
| NC score | 0.037463 (rank : 152) | θ value | 8.99809 (rank : 120) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q07954, Q2PP12, Q8IVG8 | Gene names | LRP1, A2MR | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1 precursor (LRP) (Alpha-2-macroglobulin receptor) (A2MR) (Apolipoprotein E receptor) (APOER) (CD91 antigen). | |||||
|
SIVA_MOUSE
|
||||||
| NC score | 0.035456 (rank : 153) | θ value | 1.06291 (rank : 75) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54926, Q3U6J2, Q921I8, Q9CWL1, Q9R1Q0, Q9R1Q1 | Gene names | Siva | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
TSP1_HUMAN
|
||||||
| NC score | 0.032176 (rank : 154) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
TSP1_MOUSE
|
||||||
| NC score | 0.031971 (rank : 155) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P35441 | Gene names | Thbs1, Tsp1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
SLIT3_HUMAN
|
||||||
| NC score | 0.031546 (rank : 156) | θ value | 3.0926 (rank : 95) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O75094, O95804, Q9UFH5 | Gene names | SLIT3, KIAA0814, MEGF5, SLIL2 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Multiple epidermal growth factor-like domains 5). | |||||
|
TNR6A_HUMAN
|
||||||
| NC score | 0.031177 (rank : 157) | θ value | 2.36792 (rank : 87) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NDV7, O15408, Q658L5, Q6NVB5, Q8NEZ0, Q8TBT8, Q8TCR0, Q9NV59, Q9P268 | Gene names | TNRC6A, CAGH26, KIAA1460, TNRC6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trinucleotide repeat-containing gene 6A protein (CAG repeat protein 26) (Glycine-tryptophan protein of 182 kDa) (GW182 autoantigen) (Protein GW1) (EMSY interactor protein). | |||||
|
SLIT3_MOUSE
|
||||||
| NC score | 0.029816 (rank : 158) | θ value | 8.99809 (rank : 124) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9WVB4 | Gene names | Slit3 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 3 protein precursor (Slit-3) (Slit3). | |||||
|
FGRL1_MOUSE
|
||||||
| NC score | 0.027868 (rank : 159) | θ value | 0.0252991 (rank : 47) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V87, Q920C2, Q920C3 | Gene names | Fgfrl1, Fgfr5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5). | |||||
|
TIF1B_HUMAN
|
||||||
| NC score | 0.026180 (rank : 160) | θ value | 0.813845 (rank : 73) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13263, O00677, Q7Z632, Q93040, Q96IM1 | Gene names | TRIM28, KAP1, RNF96, TIF1B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (Nuclear corepressor KAP-1) (KRAB- associated protein 1) (KAP-1) (KRAB-interacting protein 1) (KRIP-1) (RING finger protein 96). | |||||
|
TIF1B_MOUSE
|
||||||
| NC score | 0.024969 (rank : 161) | θ value | 1.06291 (rank : 77) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62318, P70391, Q8C283, Q99PN4 | Gene names | Trim28, Krip1, Tif1b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-beta (TIF1-beta) (Tripartite motif-containing protein 28) (KRAB-A-interacting protein) (KRIP-1). | |||||
|
FGRL1_HUMAN
|
||||||
| NC score | 0.024899 (rank : 162) | θ value | 0.125558 (rank : 59) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N441, Q6PJN1, Q9BXN7, Q9H4D7 | Gene names | FGFRL1, FGFR5, FHFR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibroblast growth factor receptor-like 1 precursor (FGF receptor-like protein 1) (Fibroblast growth factor receptor 5) (FGFR-like protein) (FGF homologous factor receptor). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.022064 (rank : 163) | θ value | 0.813845 (rank : 70) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.021814 (rank : 164) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
AIRE_MOUSE
|
||||||
| NC score | 0.021673 (rank : 165) | θ value | 1.06291 (rank : 74) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z0E3, Q9JLW0, Q9JLW1, Q9JLW2, Q9JLW3, Q9JLW4, Q9JLW5, Q9JLW6, Q9JLW7, Q9JLW8, Q9JLW9, Q9JLX0 | Gene names | Aire | |||
|
Domain Architecture |

|
|||||
| Description | Autoimmune regulator (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy protein homolog) (APECED protein homolog). | |||||
|
FHL3_MOUSE
|
||||||
| NC score | 0.020528 (rank : 166) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 424 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9R059, Q9JLP5, Q9WUH3 | Gene names | Fhl3 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.019872 (rank : 167) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
FHL3_HUMAN
|
||||||
| NC score | 0.018053 (rank : 168) | θ value | 5.27518 (rank : 105) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q13643 | Gene names | FHL3, SLIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 3 (FHL-3) (Skeletal muscle LIM- protein 2) (SLIM 2). | |||||
|
GP179_HUMAN
|
||||||
| NC score | 0.017539 (rank : 169) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6PRD1 | Gene names | GPR179, GPR158L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 179 precursor (Probable G-protein coupled receptor 158-like 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.016052 (rank : 170) | θ value | 0.163984 (rank : 63) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
JARD2_HUMAN
|
||||||
| NC score | 0.015860 (rank : 171) | θ value | 1.81305 (rank : 80) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92833, Q5U5L5 | Gene names | JARID2, JMJ | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
STON1_HUMAN
|
||||||
| NC score | 0.015736 (rank : 172) | θ value | 2.36792 (rank : 86) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.015148 (rank : 173) | θ value | 8.99809 (rank : 126) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
FHL2_MOUSE
|
||||||
| NC score | 0.015057 (rank : 174) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O70433, P97448 | Gene names | Fhl2 | |||
|
Domain Architecture |

|
|||||
| Description | Four and a half LIM domains protein 2 (FHL-2) (Skeletal muscle LIM- protein 3) (SLIM 3). | |||||
|
FAM5B_HUMAN
|
||||||
| NC score | 0.014888 (rank : 175) | θ value | 2.36792 (rank : 84) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9C0B6, O95560, Q6ZWC1, Q7LCZ9, Q8N360 | Gene names | FAM5B, BRINP2, DBCCR1L2, KIAA1747 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM5B precursor (BMP/retinoic acid-inducible neural-specific protein 2) (DBCCR1-like protein 2). | |||||
|
GPC6A_HUMAN
|
||||||
| NC score | 0.014529 (rank : 176) | θ value | 0.62314 (rank : 68) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5T6X5, Q6JK43, Q6JK44, Q8NGU8, Q8NHZ9, Q8TDT6 | Gene names | GPRC6A, GPCR33 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor family C group 6 member A precursor (hGPRC6A) (G-protein coupled receptor 33) (hGPCR33). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.013691 (rank : 177) | θ value | 6.88961 (rank : 110) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.012834 (rank : 178) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.012818 (rank : 179) | θ value | 6.88961 (rank : 114) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.011548 (rank : 180) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
TEX14_MOUSE
|
||||||
| NC score | 0.010882 (rank : 181) | θ value | 0.0961366 (rank : 56) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1333 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7M6U3, Q3UT36, Q5NC10, Q5NC11, Q8CGK1, Q8CGK2, Q99MV8 | Gene names | Tex14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed protein 14 (Testis-expressed sequence 14). | |||||
|
PER3_MOUSE
|
||||||
| NC score | 0.010009 (rank : 182) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70361 | Gene names | Per3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (mPER3). | |||||
|
CBL_MOUSE
|
||||||
| NC score | 0.008714 (rank : 183) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P22682, Q8CEA1 | Gene names | Cbl | |||
|
Domain Architecture |

|
|||||
| Description | E3 ubiquitin-protein ligase CBL (EC 6.3.2.-) (Signal transduction protein CBL) (Proto-oncogene c-CBL) (Casitas B-lineage lymphoma proto- oncogene). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.007828 (rank : 184) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TCF20_MOUSE
|
||||||
| NC score | 0.007545 (rank : 185) | θ value | 8.99809 (rank : 125) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EPQ8, Q60792 | Gene names | Tcf20, Spbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor 20 (Stromelysin 1 PDGF-responsive element-binding protein) (SPRE-binding protein) (Nuclear factor SPBP). | |||||
|
EPHB4_HUMAN
|
||||||
| NC score | 0.006875 (rank : 186) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1056 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P54760, Q9BTA5, Q9BXP0 | Gene names | EPHB4, HTK | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor HTK). | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.006827 (rank : 187) | θ value | 8.99809 (rank : 122) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
VINEX_MOUSE
|
||||||
| NC score | 0.006402 (rank : 188) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R1Z8, Q62423 | Gene names | Sorbs3, Scam1, Sh3d4 | |||
|
Domain Architecture |

|
|||||
| Description | Vinexin (Sorbin and SH3 domain-containing protein 3) (SH3-containing adapter molecule 1) (SCAM-1) (SH3 domain-containing protein SH3P3). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.005231 (rank : 189) | θ value | 6.88961 (rank : 113) | |||
| Query Neighborhood Hits | 127 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||