Please be patient as the page loads
|
TNR18_HUMAN
|
||||||
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TNR18_HUMAN
|
||||||
| θ value | 1.00358e-83 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
|
TNR18_MOUSE
|
||||||
| θ value | 1.18825e-76 (rank : 2) | NC score | 0.944984 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35714, Q9JKR1, Q9JKR2, Q9JKR3 | Gene names | Tnfrsf18, Gitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein). | |||||
|
TNR4_HUMAN
|
||||||
| θ value | 9.59137e-10 (rank : 3) | NC score | 0.543061 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
TNR9_HUMAN
|
||||||
| θ value | 4.76016e-09 (rank : 4) | NC score | 0.627827 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
TNR9_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 5) | NC score | 0.617541 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
TNR4_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 6) | NC score | 0.536302 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47741 | Gene names | Tnfrsf4, Ox40, Txgp1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (OX40 antigen). | |||||
|
TR11B_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 7) | NC score | 0.524675 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08712, O70202 | Gene names | Tnfrsf11b, Ocif, Opg | |||
|
Domain Architecture |
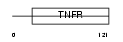
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR21_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 8) | NC score | 0.507866 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TR11B_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 9) | NC score | 0.526076 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00300, O60236, Q53FX6, Q9UHP4 | Gene names | TNFRSF11B, OCIF, OPG | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR6B_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 10) | NC score | 0.546434 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95407 | Gene names | TNFRSF6B, DCR3, TR6 | |||
|
Domain Architecture |
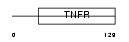
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6B precursor (Decoy receptor for Fas ligand) (Decoy receptor 3) (DcR3) (M68). | |||||
|
TNR5_MOUSE
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.518262 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |
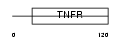
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
TNR11_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 12) | NC score | 0.525373 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |
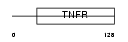
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
TNR11_MOUSE
|
||||||
| θ value | 3.19293e-05 (rank : 13) | NC score | 0.524863 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |
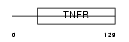
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
TNR21_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 14) | NC score | 0.487901 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR5_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 15) | NC score | 0.523648 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25942, Q9BYU0 | Gene names | CD40, TNFRSF5 | |||
|
Domain Architecture |
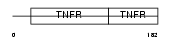
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (CDw40) (Bp50). | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.497074 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
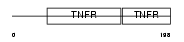
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
TNR1B_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 17) | NC score | 0.489123 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |
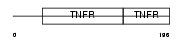
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
TNR3_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 18) | NC score | 0.472994 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
FBN2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 19) | NC score | 0.086232 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 20) | NC score | 0.085420 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
LAMB2_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.087309 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
EDAR_MOUSE
|
||||||
| θ value | 0.125558 (rank : 22) | NC score | 0.171500 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
TNR3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.473742 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P50284 | Gene names | Ltbr, Tnfcr, Tnfrsf3 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor). | |||||
|
EGFR_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.016236 (rank : 149) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||
|
TNR14_HUMAN
|
||||||
| θ value | 0.21417 (rank : 25) | NC score | 0.434050 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
TNR23_MOUSE
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.308846 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ER63, Q8VHC0 | Gene names | Tnfrsf23, Dctrailr1, Tnfrh1, Tnfrsf1al1 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 23 precursor (Tumor necrosis factor receptor p60 homolog 1) (TNF receptor family member SOB) (Decoy TRAIL receptor 1) (TNF receptor homolog 1). | |||||
|
EGFR_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.016803 (rank : 148) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
LAMC3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.089627 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LRP8_MOUSE
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.051156 (rank : 127) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
VLDLR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.054337 (rank : 107) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
FBN3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.087804 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
ITB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.054912 (rank : 101) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
LAMB2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.085774 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.032980 (rank : 145) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.033404 (rank : 144) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.084089 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
SLIT1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.036481 (rank : 143) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
ITB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 38) | NC score | 0.054060 (rank : 108) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
FBN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.083078 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.054725 (rank : 105) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
EDAR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.150882 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
ITB3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.053260 (rank : 113) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |
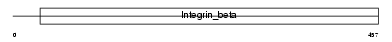
|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.047786 (rank : 140) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
CD248_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.044941 (rank : 142) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
LAMB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.068184 (rank : 67) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.071252 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SIVA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.026649 (rank : 147) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15304, Q96P98, Q9UPD6 | Gene names | SIVA | |||
|
Domain Architecture |
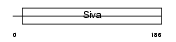
|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
TNR19_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.119745 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NS68, Q9BXZ9, Q9BY00, Q9NZV2 | Gene names | TNFRSF19, TAJ, TROY | |||
|
Domain Architecture |
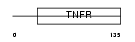
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.058917 (rank : 87) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
NFX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.045992 (rank : 141) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
ADA15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.012678 (rank : 150) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88839, Q3U7C2, Q8C7Z0, Q91VS9, Q9QYL2 | Gene names | Adam15, Mdc15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin) (AD56). | |||||
|
FSTL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.032191 (rank : 146) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.059994 (rank : 83) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
LOXL1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.009336 (rank : 151) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
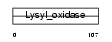
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
MGR3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.005266 (rank : 152) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.053107 (rank : 115) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
CREL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.054899 (rank : 102) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CREL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.053132 (rank : 114) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
CRIM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.050130 (rank : 139) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
CRUM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.051264 (rank : 123) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
DLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.050231 (rank : 137) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
DLL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.050855 (rank : 133) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |
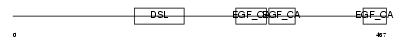
|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
FBLN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.050495 (rank : 134) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
FBLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.051087 (rank : 130) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FBLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.050192 (rank : 138) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
FRAS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.054813 (rank : 104) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
FRAS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.054056 (rank : 109) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
JAG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.051209 (rank : 125) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
JAG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.052074 (rank : 119) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.064000 (rank : 75) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA53_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.063578 (rank : 76) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051134 (rank : 129) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.053568 (rank : 110) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA56_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.064561 (rank : 74) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA57_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.055117 (rank : 99) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.061520 (rank : 80) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KRA59_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.076018 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
LAMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.061088 (rank : 81) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.062020 (rank : 79) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.057738 (rank : 92) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.052474 (rank : 118) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052689 (rank : 117) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
LAMA5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.070387 (rank : 63) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.069148 (rank : 66) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.057531 (rank : 93) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
LAMB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.054881 (rank : 103) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
LAMC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.067391 (rank : 69) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.065481 (rank : 72) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.058824 (rank : 88) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
LAMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.066806 (rank : 70) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
LAMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.074613 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
LTB1L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.055761 (rank : 98) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.057130 (rank : 94) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.055023 (rank : 100) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.057124 (rank : 95) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTBP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.058087 (rank : 91) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.058397 (rank : 90) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.059440 (rank : 86) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.060273 (rank : 82) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MEGF6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.071097 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.069521 (rank : 64) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.059819 (rank : 84) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.067659 (rank : 68) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MEGF9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.063015 (rank : 77) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
MT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.056474 (rank : 96) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28184 | Gene names | Mt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF). | |||||
|
NET4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.051181 (rank : 126) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
NOTC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.053512 (rank : 111) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
NOTC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.051262 (rank : 124) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
NOTC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.051556 (rank : 121) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.050447 (rank : 136) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
NOTC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.051150 (rank : 128) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NOTC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.051054 (rank : 131) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NTNG2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051021 (rank : 132) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4F1, Q8R4F2, Q8VIP6, Q8VIP7, Q8VIP8 | Gene names | Ntng2, Lmnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
PCSK5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.073494 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.051912 (rank : 120) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
SCUB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.071211 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SCUB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.070822 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
SREC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.062881 (rank : 78) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
SREC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.064658 (rank : 73) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
SREC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.066314 (rank : 71) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
STAB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.058494 (rank : 89) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
STAB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.059583 (rank : 85) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.055820 (rank : 97) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
STAB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.054578 (rank : 106) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
TNR16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.291459 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |
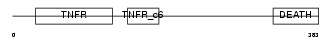
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.292960 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |
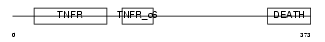
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
TNR19_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.166129 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLL3, Q812G3, Q9JHF1, Q9JJH6, Q9JLL2, Q9QXW7 | Gene names | Tnfrsf19, Taj, Troy | |||
|
Domain Architecture |
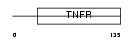
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
TNR1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.220355 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
TNR1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.237780 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.289042 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ER62, Q8VHB9, Q9CZA4 | Gene names | Tnfrsf22, Dctrailr2, Tnfrh2, Tnfrsf1al2 | |||
|
Domain Architecture |
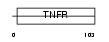
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 22 (Tumor necrosis factor receptor p60 homolog 2) (TNF receptor family member SOBa) (Decoy TRAIL receptor 2) (TNF receptor homolog 2). | |||||
|
TNR25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.221470 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TNR26_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.292955 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P83626 | Gene names | Tnfrsf26, Tnfrh3 | |||
|
Domain Architecture |
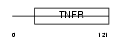
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 26 precursor (TNF receptor homolog 3). | |||||
|
TNR27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.152210 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HAV5, Q6UWM2, Q8IZA6 | Gene names | EDA2R, TNFRSF27, XEDAR | |||
|
Domain Architecture |
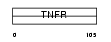
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TNR27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.121612 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BX35, Q8BM50 | Gene names | Eda2r, Tnfrsf27, Xedar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TNR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.224079 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25445, Q14292, Q14293, Q14294, Q14295, Q16652, Q6SSE9 | Gene names | FAS, APT1, FAS1, TNFRSF6 | |||
|
Domain Architecture |
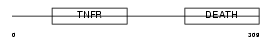
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.247158 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P25446, Q9DCQ1 | Gene names | Fas, Apt1, Tnfrsf6 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.250870 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
TNR7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.227393 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
TNR8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.261100 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TNR8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.218125 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
TR10A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.150032 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |
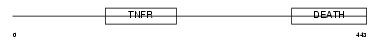
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
TR10B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.161100 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
TR10B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.136001 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZM4, Q9JJL5, Q9JJL6 | Gene names | Tnfrsf10b, Dr5, Killer | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (MK). | |||||
|
TR10C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.173266 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |
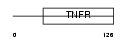
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
TR10D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.146850 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBN6, Q9Y6Q4 | Gene names | TNFRSF10D, DCR2, TRAILR4, TRUNDD | |||
|
Domain Architecture |
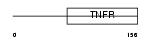
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10D precursor (Decoy receptor 2) (DcR2) (TNF-related apoptosis-inducing ligand receptor 4) (TRAIL receptor 4) (TRAIL-R4) (TRAIL receptor with a truncated death domain) (CD264 antigen). | |||||
|
TR19L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.177290 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |
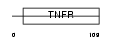
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
TR19L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.145636 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BX43, Q8BTV0 | Gene names | Tnfrsf19l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor. | |||||
|
USH2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.053440 (rank : 112) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
USH2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.053064 (rank : 116) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
WIF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.050447 (rank : 135) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
WIF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051319 (rank : 122) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
ZAN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.069221 (rank : 65) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
TNR18_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.00358e-83 (rank : 1) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
|
TNR18_MOUSE
|
||||||
| NC score | 0.944984 (rank : 2) | θ value | 1.18825e-76 (rank : 2) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O35714, Q9JKR1, Q9JKR2, Q9JKR3 | Gene names | Tnfrsf18, Gitr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein). | |||||
|
TNR9_HUMAN
|
||||||
| NC score | 0.627827 (rank : 3) | θ value | 4.76016e-09 (rank : 4) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q07011 | Gene names | TNFRSF9, CD137, ILA | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB homolog) (T-cell antigen ILA) (CD137 antigen). | |||||
|
TNR9_MOUSE
|
||||||
| NC score | 0.617541 (rank : 4) | θ value | 3.08544e-08 (rank : 5) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P20334 | Gene names | Tnfrsf9, Cd137, Cd157, Ila, Ly63 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 9 precursor (4-1BB ligand receptor) (T-cell antigen 4-1BB) (CD137 antigen). | |||||
|
TNR6B_HUMAN
|
||||||
| NC score | 0.546434 (rank : 5) | θ value | 6.43352e-06 (rank : 10) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | O95407 | Gene names | TNFRSF6B, DCR3, TR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6B precursor (Decoy receptor for Fas ligand) (Decoy receptor 3) (DcR3) (M68). | |||||
|
TNR4_HUMAN
|
||||||
| NC score | 0.543061 (rank : 6) | θ value | 9.59137e-10 (rank : 3) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P43489, Q13663, Q5T7M0 | Gene names | TNFRSF4, TXGP1L | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (ACT35 antigen) (TAX transcriptionally-activated glycoprotein 1 receptor) (CD134 antigen). | |||||
|
TNR4_MOUSE
|
||||||
| NC score | 0.536302 (rank : 7) | θ value | 5.26297e-08 (rank : 6) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P47741 | Gene names | Tnfrsf4, Ox40, Txgp1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 4 precursor (OX40L receptor) (OX40 antigen). | |||||
|
TR11B_HUMAN
|
||||||
| NC score | 0.526076 (rank : 8) | θ value | 4.92598e-06 (rank : 9) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O00300, O60236, Q53FX6, Q9UHP4 | Gene names | TNFRSF11B, OCIF, OPG | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR11_HUMAN
|
||||||
| NC score | 0.525373 (rank : 9) | θ value | 1.09739e-05 (rank : 12) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y6Q6 | Gene names | TNFRSF11A, RANK | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR) (CD265 antigen). | |||||
|
TNR11_MOUSE
|
||||||
| NC score | 0.524863 (rank : 10) | θ value | 3.19293e-05 (rank : 13) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O35305, Q8VCT7 | Gene names | Tnfrsf11a, Rank | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11A precursor (Receptor activator of NF-KB) (Osteoclast differentiation factor receptor) (ODFR). | |||||
|
TR11B_MOUSE
|
||||||
| NC score | 0.524675 (rank : 11) | θ value | 9.92553e-07 (rank : 7) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O08712, O70202 | Gene names | Tnfrsf11b, Ocif, Opg | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 11B precursor (Osteoprotegerin) (Osteoclastogenesis inhibitory factor). | |||||
|
TNR5_HUMAN
|
||||||
| NC score | 0.523648 (rank : 12) | θ value | 9.29e-05 (rank : 15) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P25942, Q9BYU0 | Gene names | CD40, TNFRSF5 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (CDw40) (Bp50). | |||||
|
TNR5_MOUSE
|
||||||
| NC score | 0.518262 (rank : 13) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P27512, Q99NE0, Q99NE1, Q99NE2, Q99NE3 | Gene names | Cd40, Tnfrsf5 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 5 precursor (CD40L receptor) (B-cell surface antigen CD40) (BP50) (CDw40). | |||||
|
TNR21_HUMAN
|
||||||
| NC score | 0.507866 (rank : 14) | θ value | 4.92598e-06 (rank : 8) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O75509, Q96D86 | Gene names | TNFRSF21, DR6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.497074 (rank : 15) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
TNR1B_HUMAN
|
||||||
| NC score | 0.489123 (rank : 16) | θ value | 0.000270298 (rank : 17) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
TNR21_MOUSE
|
||||||
| NC score | 0.487901 (rank : 17) | θ value | 5.44631e-05 (rank : 14) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9EPU5, Q91W77, Q91XH9 | Gene names | Tnfrsf21, Dr6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 21 precursor (TNFR- related death receptor 6) (Death receptor 6). | |||||
|
TNR3_MOUSE
|
||||||
| NC score | 0.473742 (rank : 18) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P50284 | Gene names | Ltbr, Tnfcr, Tnfrsf3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor). | |||||
|
TNR3_HUMAN
|
||||||
| NC score | 0.472994 (rank : 19) | θ value | 0.00175202 (rank : 18) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P36941 | Gene names | LTBR, TNFCR, TNFRSF3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 3 precursor (Lymphotoxin-beta receptor) (Tumor necrosis factor receptor 2-related protein) (Tumor necrosis factor C receptor). | |||||
|
TNR14_HUMAN
|
||||||
| NC score | 0.434050 (rank : 20) | θ value | 0.21417 (rank : 25) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92956, Q8WXR1, Q96J31, Q9UM65 | Gene names | TNFRSF14, HVEA, HVEM | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 14 precursor (Herpesvirus entry mediator A) (Tumor necrosis factor receptor-like 2) (TR2). | |||||
|
TNR23_MOUSE
|
||||||
| NC score | 0.308846 (rank : 21) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9ER63, Q8VHC0 | Gene names | Tnfrsf23, Dctrailr1, Tnfrh1, Tnfrsf1al1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 23 precursor (Tumor necrosis factor receptor p60 homolog 1) (TNF receptor family member SOB) (Decoy TRAIL receptor 1) (TNF receptor homolog 1). | |||||
|
TNR16_MOUSE
|
||||||
| NC score | 0.292960 (rank : 22) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Z0W1 | Gene names | Ngfr, Tnfrsf16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Low affinity neurotrophin receptor p75NTR). | |||||
|
TNR26_MOUSE
|
||||||
| NC score | 0.292955 (rank : 23) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P83626 | Gene names | Tnfrsf26, Tnfrh3 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 26 precursor (TNF receptor homolog 3). | |||||
|
TNR16_HUMAN
|
||||||
| NC score | 0.291459 (rank : 24) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P08138 | Gene names | NGFR, TNFRSF16 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 16 precursor (Low- affinity nerve growth factor receptor) (NGF receptor) (Gp80-LNGFR) (p75 ICD) (Low affinity neurotrophin receptor p75NTR) (CD271 antigen). | |||||
|
TNR22_MOUSE
|
||||||
| NC score | 0.289042 (rank : 25) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9ER62, Q8VHB9, Q9CZA4 | Gene names | Tnfrsf22, Dctrailr2, Tnfrh2, Tnfrsf1al2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 22 (Tumor necrosis factor receptor p60 homolog 2) (TNF receptor family member SOBa) (Decoy TRAIL receptor 2) (TNF receptor homolog 2). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.261100 (rank : 26) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
TNR7_HUMAN
|
||||||
| NC score | 0.250870 (rank : 27) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P26842 | Gene names | TNFRSF7, CD27 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27) (T14). | |||||
|
TNR6_MOUSE
|
||||||
| NC score | 0.247158 (rank : 28) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P25446, Q9DCQ1 | Gene names | Fas, Apt1, Tnfrsf6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR1A_MOUSE
|
||||||
| NC score | 0.237780 (rank : 29) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P25118 | Gene names | Tnfrsf1a, Tnfr-1, Tnfr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55). | |||||
|
TNR7_MOUSE
|
||||||
| NC score | 0.227393 (rank : 30) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P41272 | Gene names | Tnfrsf7, Cd27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 7 precursor (CD27L receptor) (T-cell activation antigen CD27). | |||||
|
TNR6_HUMAN
|
||||||
| NC score | 0.224079 (rank : 31) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P25445, Q14292, Q14293, Q14294, Q14295, Q16652, Q6SSE9 | Gene names | FAS, APT1, FAS1, TNFRSF6 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 6 precursor (FASLG receptor) (Apoptosis-mediating surface antigen FAS) (Apo-1 antigen) (CD95 antigen). | |||||
|
TNR25_HUMAN
|
||||||
| NC score | 0.221470 (rank : 32) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q93038, O00275, O00276, O00277, O00278, O00279, O00280, O14865, O14866, P78507, P78515, Q92983, Q93036, Q93037, Q99722, Q99830, Q99831, Q9BY86, Q9UME0, Q9UME1, Q9UME5 | Gene names | TNFRSF25, APO3, DDR3, DR3, TNFRSF12, WSL, WSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 25 precursor (WSL-1 protein) (Apoptosis-mediating receptor DR3) (Apoptosis-mediating receptor TRAMP) (Death domain receptor 3) (WSL protein) (Apoptosis- inducing receptor AIR) (Apo-3) (Lymphocyte-associated receptor of death) (LARD). | |||||
|
TNR1A_HUMAN
|
||||||
| NC score | 0.220355 (rank : 33) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
TNR8_MOUSE
|
||||||
| NC score | 0.218125 (rank : 34) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60846 | Gene names | Tnfrsf8 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30). | |||||
|
TR19L_HUMAN
|
||||||
| NC score | 0.177290 (rank : 35) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q969Z4, Q86V34, Q96JU1, Q9BUX7 | Gene names | TNFRSF19L, RELT | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor (Receptor expressed in lymphoid tissues). | |||||
|
TR10C_HUMAN
|
||||||
| NC score | 0.173266 (rank : 36) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O14798, O14755, Q6UXM5 | Gene names | TNFRSF10C, DCR1, LIT, TRAILR3, TRID | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10C precursor (Decoy receptor 1) (DcR1) (Decoy TRAIL receptor without death domain) (TNF- related apoptosis-inducing ligand receptor 3) (TRAIL receptor 3) (TRAIL-R3) (Trail receptor without an intracellular domain) (Lymphocyte inhibitor of TRAIL) (Antagonist decoy receptor for TRAIL/Apo-2L) (CD263 antigen). | |||||
|
EDAR_MOUSE
|
||||||
| NC score | 0.171500 (rank : 37) | θ value | 0.125558 (rank : 22) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R187, Q9DC43 | Gene names | Edar, Dl | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (Ectodermal dysplasia receptor) (Downless). | |||||
|
TNR19_MOUSE
|
||||||
| NC score | 0.166129 (rank : 38) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9JLL3, Q812G3, Q9JHF1, Q9JJH6, Q9JLL2, Q9QXW7 | Gene names | Tnfrsf19, Taj, Troy | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
TR10B_HUMAN
|
||||||
| NC score | 0.161100 (rank : 39) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14763, O14720, O15508, O15517, O15531, Q9BVE0 | Gene names | TNFRSF10B, DR5, KILLER, TRAILR2, TRICK2, ZTNFR9 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (TNF-related apoptosis-inducing ligand receptor 2) (TRAIL receptor 2) (TRAIL-R2) (CD262 antigen). | |||||
|
TNR27_HUMAN
|
||||||
| NC score | 0.152210 (rank : 40) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9HAV5, Q6UWM2, Q8IZA6 | Gene names | EDA2R, TNFRSF27, XEDAR | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
EDAR_HUMAN
|
||||||
| NC score | 0.150882 (rank : 41) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UNE0, Q9UND9 | Gene names | EDAR, DL | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member EDAR precursor (Anhidrotic ectodysplasin receptor 1) (Ectodysplasin-A receptor) (EDA- A1 receptor) (Ectodermal dysplasia receptor) (Downless homolog). | |||||
|
TR10A_HUMAN
|
||||||
| NC score | 0.150032 (rank : 42) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00220, Q96E62 | Gene names | TNFRSF10A, APO2, DR4, TRAILR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10A precursor (Death receptor 4) (TNF-related apoptosis-inducing ligand receptor 1) (TRAIL receptor 1) (TRAIL-R1) (CD261 antigen). | |||||
|
TR10D_HUMAN
|
||||||
| NC score | 0.146850 (rank : 43) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UBN6, Q9Y6Q4 | Gene names | TNFRSF10D, DCR2, TRAILR4, TRUNDD | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10D precursor (Decoy receptor 2) (DcR2) (TNF-related apoptosis-inducing ligand receptor 4) (TRAIL receptor 4) (TRAIL-R4) (TRAIL receptor with a truncated death domain) (CD264 antigen). | |||||
|
TR19L_MOUSE
|
||||||
| NC score | 0.145636 (rank : 44) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BX43, Q8BTV0 | Gene names | Tnfrsf19l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 19L precursor. | |||||
|
TR10B_MOUSE
|
||||||
| NC score | 0.136001 (rank : 45) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QZM4, Q9JJL5, Q9JJL6 | Gene names | Tnfrsf10b, Dr5, Killer | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 10B precursor (Death receptor 5) (MK). | |||||
|
TNR27_MOUSE
|
||||||
| NC score | 0.121612 (rank : 46) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BX35, Q8BM50 | Gene names | Eda2r, Tnfrsf27, Xedar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 27 (X-linked ectodysplasin-A2 receptor) (EDA-A2 receptor). | |||||
|
TNR19_HUMAN
|
||||||
| NC score | 0.119745 (rank : 47) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NS68, Q9BXZ9, Q9BY00, Q9NZV2 | Gene names | TNFRSF19, TAJ, TROY | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 19 precursor (Toxicity and JNK inducer) (TRADE). | |||||
|
LAMC3_MOUSE
|
||||||
| NC score | 0.089627 (rank : 48) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
FBN3_HUMAN
|
||||||
| NC score | 0.087804 (rank : 49) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q75N90, Q75N91, Q75N92, Q75N93, Q86SJ5, Q96JP8 | Gene names | FBN3, KIAA1776 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibrillin-3 precursor. | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.087309 (rank : 50) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
FBN2_MOUSE
|
||||||
| NC score | 0.086232 (rank : 51) | θ value | 0.0193708 (rank : 19) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q61555, Q63957 | Gene names | Fbn2, Fbn-2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
LAMB2_HUMAN
|
||||||
| NC score | 0.085774 (rank : 52) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P55268, Q16321 | Gene names | LAMB2, LAMS | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (Laminin B1s chain). | |||||
|
FBN2_HUMAN
|
||||||
| NC score | 0.085420 (rank : 53) | θ value | 0.0431538 (rank : 20) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P35556 | Gene names | FBN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-2 precursor. | |||||
|
FBN1_HUMAN
|
||||||
| NC score | 0.084089 (rank : 54) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P35555, Q15972, Q75N87 | Gene names | FBN1, FBN | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
FBN1_MOUSE
|
||||||
| NC score | 0.083078 (rank : 55) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 563 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q61554, Q60826 | Gene names | Fbn1, Fbn-1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibrillin-1 precursor. | |||||
|
KRA59_HUMAN
|
||||||
| NC score | 0.076018 (rank : 56) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P26371, Q14564 | Gene names | KRTAP5-9, KAP5.9, KRN1, KRTAP5.9, UHSK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-9 (Keratin-associated protein 5.9) (Ultrahigh sulfur keratin-associated protein 5.9) (Keratin, cuticle, ultrahigh sulfur 1) (Keratin, ultra high-sulfur matrix protein A) (UHS keratin A) (UHS KerA). | |||||
|
LAMC3_HUMAN
|
||||||
| NC score | 0.074613 (rank : 57) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Y6N6 | Gene names | LAMC3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
PCSK5_MOUSE
|
||||||
| NC score | 0.073494 (rank : 58) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q04592, Q62040 | Gene names | Pcsk5 | |||
|
Domain Architecture |

|
|||||
| Description | Proprotein convertase subtilisin/kexin type 5 precursor (EC 3.4.21.-) (Proprotein convertase PC5) (Subtilisin/kexin-like protease PC5) (PC6) (Subtilisin-like proprotein convertase 6) (SPC6). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.071252 (rank : 59) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
SCUB3_HUMAN
|
||||||
| NC score | 0.071211 (rank : 60) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8IX30, Q5CZB3, Q86UZ9, Q8NAU9 | Gene names | SCUBE3, CEGF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
MEGF6_HUMAN
|
||||||
| NC score | 0.071097 (rank : 61) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O75095, Q5VV39 | Gene names | MEGF6, EGFL3, KIAA0815 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple epidermal growth factor-like domains 6 precursor (EGF-like domain-containing protein 3) (Multiple EGF-like domain protein 3). | |||||
|
SCUB3_MOUSE
|
||||||
| NC score | 0.070822 (rank : 62) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q66PY1, Q68FG9 | Gene names | Scube3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal peptide, CUB and EGF-like domain-containing protein 3 precursor. | |||||
|
LAMA5_HUMAN
|
||||||
| NC score | 0.070387 (rank : 63) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | O15230, Q8WZA7, Q9H1P1 | Gene names | LAMA5, KIAA0533, KIAA1907 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.069521 (rank : 64) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
ZAN_MOUSE
|
||||||
| NC score | 0.069221 (rank : 65) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 808 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O88799, O08647 | Gene names | Zan | |||
|
Domain Architecture |

|
|||||
| Description | Zonadhesin precursor. | |||||
|
LAMA5_MOUSE
|
||||||
| NC score | 0.069148 (rank : 66) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q61001, Q9JHQ6 | Gene names | Lama5 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-5 chain precursor. | |||||
|
LAMB1_HUMAN
|
||||||
| NC score | 0.068184 (rank : 67) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1018 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P07942 | Gene names | LAMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MEGF9_HUMAN
|
||||||
| NC score | 0.067659 (rank : 68) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 557 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9H1U4, O75098 | Gene names | MEGF9, EGFL5, KIAA0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
LAMC1_HUMAN
|
||||||
| NC score | 0.067391 (rank : 69) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 922 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11047 | Gene names | LAMC1, LAMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
LAMC2_MOUSE
|
||||||
| NC score | 0.066806 (rank : 70) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q61092 | Gene names | Lamc2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain). | |||||
|
SREC_HUMAN
|
||||||
| NC score | 0.066314 (rank : 71) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14162, O43701 | Gene names | SCARF1, KIAA0149, SREC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endothelial cells scavenger receptor precursor (Acetyl LDL receptor) (Scavenger receptor class F member 1). | |||||
|
LAMC1_MOUSE
|
||||||
| NC score | 0.065481 (rank : 72) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P02468 | Gene names | Lamc1, Lamb-2, Lamc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-1 chain precursor (Laminin B2 chain). | |||||
|
SREC2_MOUSE
|
||||||
| NC score | 0.064658 (rank : 73) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 605 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P59222 | Gene names | Scarf2, Srec2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II). | |||||
|
KRA56_HUMAN
|
||||||
| NC score | 0.064561 (rank : 74) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q6L8G9 | Gene names | KRTAP5-6, KAP5.6, KRTAP5.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-6 (Keratin-associated protein 5.6) (Ultrahigh sulfur keratin-associated protein 5.6). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.064000 (rank : 75) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KRA53_HUMAN
|
||||||
| NC score | 0.063578 (rank : 76) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6L8H2, Q6PL44, Q701N3 | Gene names | KRTAP5-3, KAP5-9, KAP5.3, KRTAP5-9, KRTAP5.3, KRTAP5.9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-3 (Keratin-associated protein 5.3) (Ultrahigh sulfur keratin-associated protein 5.3) (Keratin-associated protein 5-9) (Keratin-associated protein 5.9) (UHS KerB-like). | |||||
|
MEGF9_MOUSE
|
||||||
| NC score | 0.063015 (rank : 77) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BH27, Q8BWI4 | Gene names | Megf9, Egfl5, Kiaa0818 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 9 precursor (EGF-like domain-containing protein 5) (Multiple EGF-like domain protein 5). | |||||
|
SREC2_HUMAN
|
||||||
| NC score | 0.062881 (rank : 78) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96GP6, Q8IXF3, Q9BW74 | Gene names | SCARF2, SREC2, SREPCR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class F member 2 precursor (Scavenger receptor expressed by endothelial cells 2 protein) (SREC-II) (SRECRP-1). | |||||
|
LAMA1_MOUSE
|
||||||
| NC score | 0.062020 (rank : 79) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P19137 | Gene names | Lama1, Lama, Lama-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.061520 (rank : 80) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
LAMA1_HUMAN
|
||||||
| NC score | 0.061088 (rank : 81) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 804 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P25391 | Gene names | LAMA1, LAMA | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-1 chain precursor (Laminin A chain). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.060273 (rank : 82) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.059994 (rank : 83) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.059819 (rank : 84) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
STAB1_MOUSE
|
||||||
| NC score | 0.059583 (rank : 85) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8R4Y4, Q8K0K6, Q8VC09 | Gene names | Stab1, Feel1 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein). | |||||
|
LTBP3_HUMAN
|
||||||
| NC score | 0.059440 (rank : 86) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NS15, O15107, Q96HB9, Q9H7K2, Q9UFN4 | Gene names | LTBP3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.058917 (rank : 87) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
LAMC2_HUMAN
|
||||||
| NC score | 0.058824 (rank : 88) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q13753, Q02536, Q02537, Q13752, Q14941, Q5VYE8 | Gene names | LAMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-2 chain precursor (Laminin 5 gamma 2 subunit) (Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t chain) (Cell- scattering factor 140 kDa subunit) (CSF 140 kDa subunit) (Large adhesive scatter factor 140 kDa subunit) (Ladsin 140 kDa subunit). | |||||
|
STAB1_HUMAN
|
||||||
| NC score | 0.058494 (rank : 89) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 546 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9NY15, Q8IUH0, Q8IUH1, Q93072 | Gene names | STAB1, FEEL1, KIAA0246 | |||
|
Domain Architecture |

|
|||||
| Description | Stabilin-1 precursor (FEEL-1 protein) (MS-1 antigen). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.058397 (rank : 90) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LTBP2_HUMAN
|
||||||
| NC score | 0.058087 (rank : 91) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 552 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14767, Q99907, Q9NS51 | Gene names | LTBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.057738 (rank : 92) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
LAMB3_HUMAN
|
||||||
| NC score | 0.057531 (rank : 93) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13751, O14947, Q14733, Q9UJK4, Q9UJL1 | Gene names | LAMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Laminin B1k chain) (Kalinin B1 chain). | |||||
|
LTB1L_MOUSE
|
||||||
| NC score | 0.057130 (rank : 94) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8CG19, O88349, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
LTB1S_MOUSE
|
||||||
| NC score | 0.057124 (rank : 95) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CG18, Q8BNW7, Q8C7F5, Q8CIR0 | Gene names | Ltbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
MT3_MOUSE
|
||||||
| NC score | 0.056474 (rank : 96) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28184 | Gene names | Mt3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metallothionein-3 (MT-3) (Metallothionein-III) (MT-III) (Growth inhibitory factor) (GIF). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.055820 (rank : 97) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
LTB1L_HUMAN
|
||||||
| NC score | 0.055761 (rank : 98) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14766 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1L precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
KRA57_HUMAN
|
||||||
| NC score | 0.055117 (rank : 99) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6L8G8, Q701N5 | Gene names | KRTAP5-7, KAP5-7, KAP5.3, KRTAP5.3, KRTAP5.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-7 (Keratin-associated protein 5.7) (Ultrahigh sulfur keratin-associated protein 5.7) (Keratin-associated protein 5-3) (Keratin-associated protein 5.3). | |||||
|
LTB1S_HUMAN
|
||||||
| NC score | 0.055023 (rank : 100) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P22064, Q8TD95 | Gene names | LTBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein, isoform 1S precursor (LTBP-1) (Transforming growth factor beta-1-binding protein 1) (TGF-beta1-BP-1). | |||||
|
ITB2_HUMAN
|
||||||
| NC score | 0.054912 (rank : 101) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P05107, Q16418 | Gene names | ITGB2, CD18 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-2 precursor (Cell surface adhesion glycoproteins LFA- 1/CR3/p150,95 subunit beta) (Complement receptor C3 subunit beta) (CD18 antigen). | |||||
|
CREL1_HUMAN
|
||||||
| NC score | 0.054899 (rank : 102) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96HD1, Q6I9X5, Q8NFT4, Q9Y409 | Gene names | CRELD1, CIRRIN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
LAMB3_MOUSE
|
||||||
| NC score | 0.054881 (rank : 103) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q61087 | Gene names | Lamb3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-3 chain precursor (Laminin 5 beta 3) (Kalinin B1 chain). | |||||
|
FRAS1_HUMAN
|
||||||
| NC score | 0.054813 (rank : 104) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 759 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q86XX4, Q86UZ4, Q8N3U9, Q8NAU7, Q96JW7, Q9H6N9, Q9P228 | Gene names | FRAS1, KIAA1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.054725 (rank : 105) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
STAB2_MOUSE
|
||||||
| NC score | 0.054578 (rank : 106) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8R4U0, Q8BM87 | Gene names | Stab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor [Contains: Short form stabilin-2]. | |||||
|
VLDLR_HUMAN
|
||||||
| NC score | 0.054337 (rank : 107) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P98155, Q5VVF6 | Gene names | VLDLR | |||
|
Domain Architecture |

|
|||||
| Description | Very low-density lipoprotein receptor precursor (VLDL receptor) (VLDL- R). | |||||
|
ITB3_HUMAN
|
||||||
| NC score | 0.054060 (rank : 108) | θ value | 1.81305 (rank : 38) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P05106, O15495, Q13413, Q14648, Q16499 | Gene names | ITGB3, GP3A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
FRAS1_MOUSE
|
||||||
| NC score | 0.054056 (rank : 109) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q80T14, Q80TC7, Q811H8, Q8BPZ4 | Gene names | Fras1, Kiaa1500 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Extracellular matrix protein FRAS1 precursor. | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.053568 (rank : 110) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
NOTC1_HUMAN
|
||||||
| NC score | 0.053512 (rank : 111) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P46531 | Gene names | NOTCH1, TAN1 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (hN1) (Translocation-associated notch protein TAN-1) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
USH2A_HUMAN
|
||||||
| NC score | 0.053440 (rank : 112) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | O75445, Q5VVM9, Q6S362, Q9NS27 | Gene names | USH2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein) (Usher syndrome type IIa protein). | |||||
|
ITB3_MOUSE
|
||||||
| NC score | 0.053260 (rank : 113) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O54890 | Gene names | Itgb3 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-3 precursor (Platelet membrane glycoprotein IIIa) (GPIIIa) (CD61 antigen). | |||||
|
CREL1_MOUSE
|
||||||
| NC score | 0.053132 (rank : 114) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 524 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q91XD7, Q8BGJ8 | Gene names | Creld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich with EGF-like domain protein 1 precursor. | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.053107 (rank : 115) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
USH2A_MOUSE
|
||||||
| NC score | 0.053064 (rank : 116) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q2QI47, Q9JLP3 | Gene names | Ush2A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Usherin precursor (Usher syndrome type-2A protein homolog) (Usher syndrome type IIa protein homolog). | |||||
|
LAMA3_HUMAN
|
||||||
| NC score | 0.052689 (rank : 117) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 760 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q16787, Q13679, Q13680, Q96TG0 | Gene names | LAMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Epiligrin 170 kDa subunit) (E170) (Nicein subunit alpha). | |||||
|
LAMA2_MOUSE
|
||||||
| NC score | 0.052474 (rank : 118) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1198 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q60675, Q05003, Q64061 | Gene names | Lama2 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
JAG1_MOUSE
|
||||||
| NC score | 0.052074 (rank : 119) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QXX0 | Gene names | Jag1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (CD339 antigen). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.051912 (rank : 120) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
NOTC3_HUMAN
|
||||||
| NC score | 0.051556 (rank : 121) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1062 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UM47, Q9UEB3, Q9UPL3, Q9Y6L8 | Gene names | NOTCH3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
WIF1_MOUSE
|
||||||
| NC score | 0.051319 (rank : 122) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9WUA1 | Gene names | Wif1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
CRUM1_MOUSE
|
||||||
| NC score | 0.051264 (rank : 123) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8VHS2, Q6ST50, Q71JF2, Q8BGR4 | Gene names | Crb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Crumbs homolog 1 precursor. | |||||
|
NOTC2_HUMAN
|
||||||
| NC score | 0.051262 (rank : 124) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q04721, Q99734, Q9H240 | Gene names | NOTCH2 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 2 precursor (Notch 2) (hN2) [Contains: Notch 2 extracellular truncation; Notch 2 intracellular domain]. | |||||
|
JAG1_HUMAN
|
||||||
| NC score | 0.051209 (rank : 125) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 514 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P78504, O14902, O15122, Q15816 | Gene names | JAG1, JAGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-1 precursor (Jagged1) (hJ1) (CD339 antigen). | |||||
|
NET4_HUMAN
|
||||||
| NC score | 0.051181 (rank : 126) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9HB63, Q658K9, Q7L3F1, Q7L9D6, Q7Z5B6, Q9BZP1, Q9NT44, Q9P133 | Gene names | NTN4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Netrin-4 precursor (Beta-netrin) (Hepar-derived netrin-like protein). | |||||
|
LRP8_MOUSE
|
||||||
| NC score | 0.051156 (rank : 127) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q924X6, Q8CAK9, Q8CDF5, Q921B6 | Gene names | Lrp8, Apoer2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
NOTC4_HUMAN
|
||||||
| NC score | 0.051150 (rank : 128) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 905 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q99466, O00306, Q99458, Q99940, Q9H3S8, Q9UII9, Q9UIJ0 | Gene names | NOTCH4 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) (hNotch4) [Contains: Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.051134 (rank : 129) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
FBLN2_HUMAN
|
||||||
| NC score | 0.051087 (rank : 130) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P98095 | Gene names | FBLN2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
NOTC4_MOUSE
|
||||||
| NC score | 0.051054 (rank : 131) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P31695, O35442, O88314, O88316, Q62389, Q62390, Q9R1W9, Q9R1X0 | Gene names | Notch4, Int-3, Int3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 4 precursor (Notch 4) [Contains: Transforming protein Int-3; Notch 4 extracellular truncation; Notch 4 intracellular domain]. | |||||
|
NTNG2_MOUSE
|
||||||
| NC score | 0.051021 (rank : 132) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8R4F1, Q8R4F2, Q8VIP6, Q8VIP7, Q8VIP8 | Gene names | Ntng2, Lmnt2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
DLL4_HUMAN
|
||||||
| NC score | 0.050855 (rank : 133) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NR61, Q9NQT9 | Gene names | DLL4 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 4 precursor (Drosophila Delta homolog 4). | |||||
|
FBLN1_MOUSE
|
||||||
| NC score | 0.050495 (rank : 134) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q08879, Q08878, Q8C3B1, Q91ZC9, Q922K8 | Gene names | Fbln1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-1 precursor (Basement-membrane protein 90) (BM-90). | |||||
|
WIF1_HUMAN
|
||||||
| NC score | 0.050447 (rank : 135) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y5W5, Q6UXI1, Q8WVG4 | Gene names | WIF1 | |||
|
Domain Architecture |

|
|||||
| Description | Wnt inhibitory factor 1 precursor (WIF-1). | |||||
|
NOTC3_MOUSE
|
||||||
| NC score | 0.050447 (rank : 136) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q61982 | Gene names | Notch3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 3 precursor (Notch 3) [Contains: Notch 3 extracellular truncation; Notch 3 intracellular domain]. | |||||
|
DLL1_MOUSE
|
||||||
| NC score | 0.050231 (rank : 137) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q61483 | Gene names | Dll1 | |||
|
Domain Architecture |

|
|||||
| Description | Delta-like protein 1 precursor (Drosophila Delta homolog 1) (Delta1). | |||||
|
FBLN2_MOUSE
|
||||||
| NC score | 0.050192 (rank : 138) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P37889, Q9WUI2 | Gene names | Fbln2 | |||
|
Domain Architecture |

|
|||||
| Description | Fibulin-2 precursor. | |||||
|
CRIM1_HUMAN
|
||||||
| NC score | 0.050130 (rank : 139) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZV1, Q59GH0, Q7LCQ5, Q9H318 | Gene names | CRIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich motor neuron 1 protein precursor (CRIM-1) (Cysteine-rich repeat-containing protein S52). | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.047786 (rank : 140) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
NFX1_HUMAN
|
||||||
| NC score | 0.045992 (rank : 141) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q12986 | Gene names | NFX1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor NF-X1 (EC 6.3.2.-) (Nuclear transcription factor, X box-binding, 1). | |||||
|
CD248_HUMAN
|
||||||
| NC score | 0.044941 (rank : 142) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 679 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCU0, Q3SX55, Q96KB6 | Gene names | CD248, CD164L1, TEM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Endosialin precursor (Tumor endothelial marker 1) (CD248 antigen). | |||||
|
SLIT1_HUMAN
|
||||||
| NC score | 0.036481 (rank : 143) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 700 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O75093, Q8WWZ2, Q9UIL7 | Gene names | SLIT1, KIAA0813, MEGF4, SLIL1 | |||
|
Domain Architecture |

|
|||||
| Description | Slit homolog 1 protein precursor (Slit-1) (Multiple epidermal growth factor-like domains 4). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.033404 (rank : 144) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.032980 (rank : 145) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
FSTL3_HUMAN
|
||||||
| NC score | 0.032191 (rank : 146) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95633 | Gene names | FSTL3, FLRG | |||
|
Domain Architecture |

|
|||||
| Description | Follistatin-related protein 3 precursor (Follistatin-like 3) (Follistatin-related gene protein). | |||||
|
SIVA_HUMAN
|
||||||
| NC score | 0.026649 (rank : 147) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15304, Q96P98, Q9UPD6 | Gene names | SIVA | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulatory protein Siva (CD27-binding protein) (CD27BP). | |||||
|
EGFR_HUMAN
|
||||||
| NC score | 0.016803 (rank : 148) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P00533, O00688, O00732, P06268, Q14225, Q92795, Q9BZS2, Q9GZX1, Q9H2C9, Q9H3C9, Q9UMD7, Q9UMD8, Q9UMG5 | Gene names | EGFR, ERBB1 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1) (Receptor tyrosine-protein kinase ErbB-1). | |||||
|
EGFR_MOUSE
|
||||||
| NC score | 0.016236 (rank : 149) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 927 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01279 | Gene names | Egfr | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor precursor (EC 2.7.10.1). | |||||
|
ADA15_MOUSE
|
||||||
| NC score | 0.012678 (rank : 150) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88839, Q3U7C2, Q8C7Z0, Q91VS9, Q9QYL2 | Gene names | Adam15, Mdc15 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 15 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 15) (Metalloproteinase-like, disintegrin-like, and cysteine- rich protein 15) (MDC-15) (Metalloprotease RGD disintegrin protein) (Metargidin) (AD56). | |||||
|
LOXL1_MOUSE
|
||||||
| NC score | 0.009336 (rank : 151) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
MGR3_HUMAN
|
||||||
| NC score | 0.005266 (rank : 152) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 55 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14832, Q75MV4, Q75N17, Q86YG6, Q8TBH9 | Gene names | GRM3, GPRC1C, MGLUR3 | |||
|
Domain Architecture |

|
|||||
| Description | Metabotropic glutamate receptor 3 precursor (mGluR3). | |||||