Please be patient as the page loads
|
LOXL1_MOUSE
|
||||||
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LOXL1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.991244 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
LYOX_MOUSE
|
||||||
| θ value | 1.88391e-98 (rank : 3) | NC score | 0.970095 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LYOX_HUMAN
|
||||||
| θ value | 1.22112e-97 (rank : 4) | NC score | 0.972109 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LOXL3_HUMAN
|
||||||
| θ value | 9.45883e-58 (rank : 5) | NC score | 0.719512 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL3_MOUSE
|
||||||
| θ value | 9.45883e-58 (rank : 6) | NC score | 0.719263 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 3.974e-56 (rank : 7) | NC score | 0.716424 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL4_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 8) | NC score | 0.712985 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL4_MOUSE
|
||||||
| θ value | 9.78833e-55 (rank : 9) | NC score | 0.713327 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 1.08223e-53 (rank : 10) | NC score | 0.712529 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.057652 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
BCAR1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 12) | NC score | 0.051552 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.021259 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
BCAR1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.041267 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.025531 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.014533 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.021664 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
SVIL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.017752 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.022824 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
FOXF1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.008672 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12946, Q5FWE5 | Gene names | FOXF1, FKHL5, FREAC1 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Forkhead-related activator 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.020255 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ETV5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.026474 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
BSN_HUMAN
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.017490 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
HXD3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.005181 (rank : 71) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.016918 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.007549 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
MN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.028860 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.021307 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
ZO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.011464 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.011247 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
APBA3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.012711 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
CJ046_MOUSE
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.029368 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0X2, Q3TE79, Q9CY95 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46 homolog. | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.007628 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
NDUA8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.022927 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51970, Q9Y6N0 | Gene names | NDUFA8 | |||
|
Domain Architecture |
|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 19 kDa subunit) (Complex I-19kD) (CI-19kD) (Complex I-PGIV) (CI-PGIV). | |||||
|
SI1L1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.009877 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
WASL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.022306 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
ADA12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.004844 (rank : 72) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.027360 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.027414 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.007699 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.015255 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TACT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.019598 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40200 | Gene names | CD96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell surface protein tactile precursor (T cell-activated increased late expression protein) (CD96 antigen). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.037253 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
DEN2A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.007492 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
EP300_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.014297 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
PRRT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.019833 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
CXX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.022603 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15255 | Gene names | CXX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX box protein 1 (Cerebral protein 5). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.001597 (rank : 75) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
RC3H1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.010108 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
VGFR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.001877 (rank : 74) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35969, O55094, Q61517 | Gene names | Flt1, Emrk2, Flt | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 1 precursor (EC 2.7.10.1) (VEGFR-1) (Tyrosine-protein kinase receptor FLT) (FLT-1) (Embryonic receptor kinase 2). | |||||
|
AFAD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.006055 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.013717 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.008732 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
EGFL9_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.000891 (rank : 76) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.006146 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TNR18_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.009336 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
|
TS13_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.012560 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
VSX1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.002245 (rank : 73) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZR4, Q5TF40, Q5TF41, Q9HCU3, Q9NU27 | Gene names | VSX1, RINX | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
C163A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.263203 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.265462 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.264283 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
CD5L_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.286341 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.282959 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
CD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.077592 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |
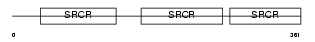
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
CD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.077561 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |
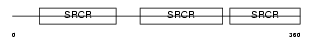
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
CD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.266159 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
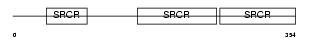
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.260222 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |
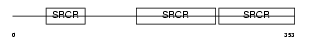
|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.220039 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.202788 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
LG3BP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.274284 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
MARCO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.171709 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.134233 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
MSRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.196034 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
MSRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.213936 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
NETR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.136607 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.136648 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
LOXL1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
LOXL1_HUMAN
|
||||||
| NC score | 0.991244 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LYOX_HUMAN
|
||||||
| NC score | 0.972109 (rank : 3) | θ value | 1.22112e-97 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| NC score | 0.970095 (rank : 4) | θ value | 1.88391e-98 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LOXL3_HUMAN
|
||||||
| NC score | 0.719512 (rank : 5) | θ value | 9.45883e-58 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
LOXL3_MOUSE
|
||||||
| NC score | 0.719263 (rank : 6) | θ value | 9.45883e-58 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.716424 (rank : 7) | θ value | 3.974e-56 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL4_MOUSE
|
||||||
| NC score | 0.713327 (rank : 8) | θ value | 9.78833e-55 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL4_HUMAN
|
||||||
| NC score | 0.712985 (rank : 9) | θ value | 3.36421e-55 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.712529 (rank : 10) | θ value | 1.08223e-53 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
CD5L_HUMAN
|
||||||
| NC score | 0.286341 (rank : 11) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| NC score | 0.282959 (rank : 12) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
LG3BP_HUMAN
|
||||||
| NC score | 0.274284 (rank : 13) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.266159 (rank : 14) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
C163A_MOUSE
|
||||||
| NC score | 0.265462 (rank : 15) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
C163B_HUMAN
|
||||||
| NC score | 0.264283 (rank : 16) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
C163A_HUMAN
|
||||||
| NC score | 0.263203 (rank : 17) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
CD6_MOUSE
|
||||||
| NC score | 0.260222 (rank : 18) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.220039 (rank : 19) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
MSRE_MOUSE
|
||||||
| NC score | 0.213936 (rank : 20) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.202788 (rank : 21) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MSRE_HUMAN
|
||||||
| NC score | 0.196034 (rank : 22) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
MARCO_HUMAN
|
||||||
| NC score | 0.171709 (rank : 23) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.136648 (rank : 24) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.136607 (rank : 25) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.134233 (rank : 26) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
CD5_HUMAN
|
||||||
| NC score | 0.077592 (rank : 27) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
CD5_MOUSE
|
||||||
| NC score | 0.077561 (rank : 28) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.057652 (rank : 29) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
BCAR1_HUMAN
|
||||||
| NC score | 0.051552 (rank : 30) | θ value | 0.0252991 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P56945 | Gene names | BCAR1, CAS, CRKAS | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
BCAR1_MOUSE
|
||||||
| NC score | 0.041267 (rank : 31) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61140, Q60869 | Gene names | Bcar1, Cas, Crkas | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer anti-estrogen resistance protein 1 (CRK-associated substrate) (p130cas). | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.037253 (rank : 32) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
CJ046_MOUSE
|
||||||
| NC score | 0.029368 (rank : 33) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0X2, Q3TE79, Q9CY95 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf46 homolog. | |||||
|
MN1_HUMAN
|
||||||
| NC score | 0.028860 (rank : 34) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q10571 | Gene names | MN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable tumor suppressor protein MN1. | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.027414 (rank : 35) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.027360 (rank : 36) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
ETV5_MOUSE
|
||||||
| NC score | 0.026474 (rank : 37) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CXC9, Q3TG49, Q8C0F3, Q9JHB1 | Gene names | Etv5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 5. | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.025531 (rank : 38) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
NDUA8_HUMAN
|
||||||
| NC score | 0.022927 (rank : 39) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51970, Q9Y6N0 | Gene names | NDUFA8 | |||
|
Domain Architecture |

|
|||||
| Description | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 (EC 1.6.5.3) (EC 1.6.99.3) (NADH-ubiquinone oxidoreductase 19 kDa subunit) (Complex I-19kD) (CI-19kD) (Complex I-PGIV) (CI-PGIV). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.022824 (rank : 40) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CXX1_HUMAN
|
||||||
| NC score | 0.022603 (rank : 41) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O15255 | Gene names | CXX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CAAX box protein 1 (Cerebral protein 5). | |||||
|
WASL_HUMAN
|
||||||
| NC score | 0.022306 (rank : 42) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00401, Q7Z746 | Gene names | WASL | |||
|
Domain Architecture |

|
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.021664 (rank : 43) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.021307 (rank : 44) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.021259 (rank : 45) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.020255 (rank : 46) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PRRT1_HUMAN
|
||||||
| NC score | 0.019833 (rank : 47) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99946, Q96DW3, Q96NQ8 | Gene names | PRRT1, C6orf31, NG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 1. | |||||
|
TACT_HUMAN
|
||||||
| NC score | 0.019598 (rank : 48) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40200 | Gene names | CD96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell surface protein tactile precursor (T cell-activated increased late expression protein) (CD96 antigen). | |||||
|
SVIL_MOUSE
|
||||||
| NC score | 0.017752 (rank : 49) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K4L3 | Gene names | Svil | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Supervillin (Archvillin) (p205/p250). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.017490 (rank : 50) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.016918 (rank : 51) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.015255 (rank : 52) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.014533 (rank : 53) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
EP300_HUMAN
|
||||||
| NC score | 0.014297 (rank : 54) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q09472 | Gene names | EP300, P300 | |||
|
Domain Architecture |

|
|||||
| Description | E1A-associated protein p300 (EC 2.3.1.48). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.013717 (rank : 55) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
APBA3_HUMAN
|
||||||
| NC score | 0.012711 (rank : 56) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O96018, O60483 | Gene names | APBA3, MINT3, X11L2 | |||
|
Domain Architecture |

|
|||||
| Description | Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma). | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.012560 (rank : 57) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
ZO1_HUMAN
|
||||||
| NC score | 0.011464 (rank : 58) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q07157, Q4ZGJ6 | Gene names | TJP1, ZO1 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-1 (Zonula occludens 1 protein) (Zona occludens 1 protein) (Tight junction protein 1). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.011247 (rank : 59) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
RC3H1_HUMAN
|
||||||
| NC score | 0.010108 (rank : 60) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5TC82, Q5W180, Q5W181, Q8IVE6, Q8N9V1 | Gene names | RC3H1, KIAA2025 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Roquin (RING finger and C3H zinc finger protein 1). | |||||
|
SI1L1_MOUSE
|
||||||
| NC score | 0.009877 (rank : 61) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C0T5, Q6PDI8, Q80U02, Q8C026 | Gene names | Sipa1l1, Kiaa0440 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal-induced proliferation-associated 1-like protein 1. | |||||
|
TNR18_HUMAN
|
||||||
| NC score | 0.009336 (rank : 62) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5U5, O95851, Q9NYJ9 | Gene names | TNFRSF18, AITR, GITR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor necrosis factor receptor superfamily member 18 precursor (Glucocorticoid-induced TNFR-related protein) (Activation-inducible TNFR family receptor). | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.008732 (rank : 63) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
FOXF1_HUMAN
|
||||||
| NC score | 0.008672 (rank : 64) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q12946, Q5FWE5 | Gene names | FOXF1, FKHL5, FREAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein F1 (Forkhead-related protein FKHL5) (Forkhead- related transcription factor 1) (FREAC-1) (Forkhead-related activator 1). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.007699 (rank : 65) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.007628 (rank : 66) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.007549 (rank : 67) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
DEN2A_HUMAN
|
||||||
| NC score | 0.007492 (rank : 68) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.006146 (rank : 69) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
AFAD_HUMAN
|
||||||
| NC score | 0.006055 (rank : 70) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P55196, O75087, O75088, O75089, Q5TIG6, Q9NSN7, Q9NU92 | Gene names | MLLT4, AF6 | |||
|
Domain Architecture |

|
|||||
| Description | Afadin (Protein AF-6). | |||||
|
HXD3_MOUSE
|
||||||
| NC score | 0.005181 (rank : 71) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09027, Q3UUD3 | Gene names | Hoxd3, Hox-4.1, Hoxd-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D3 (Hox-4.1) (Homeobox protein MH-19). | |||||
|
ADA12_MOUSE
|
||||||
| NC score | 0.004844 (rank : 72) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61824 | Gene names | Adam12, Mltna | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 12 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 12) (Meltrin alpha). | |||||
|
VSX1_HUMAN
|
||||||
| NC score | 0.002245 (rank : 73) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NZR4, Q5TF40, Q5TF41, Q9HCU3, Q9NU27 | Gene names | VSX1, RINX | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
VGFR1_MOUSE
|
||||||
| NC score | 0.001877 (rank : 74) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1118 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35969, O55094, Q61517 | Gene names | Flt1, Emrk2, Flt | |||
|
Domain Architecture |

|
|||||
| Description | Vascular endothelial growth factor receptor 1 precursor (EC 2.7.10.1) (VEGFR-1) (Tyrosine-protein kinase receptor FLT) (FLT-1) (Embryonic receptor kinase 2). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.001597 (rank : 75) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
EGFL9_HUMAN
|
||||||
| NC score | 0.000891 (rank : 76) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 365 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UY11, Q9BQ54 | Gene names | EGFL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EGF-like domain-containing protein 9 precursor (Multiple EGF-like domain protein 9). | |||||