Please be patient as the page loads
|
LG3BP_HUMAN
|
||||||
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |
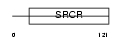
|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LG3BP_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
C163A_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 2) | NC score | 0.854556 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
DMBT1_HUMAN
|
||||||
| θ value | 2.59387e-31 (rank : 3) | NC score | 0.685159 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
DMBT1_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 4) | NC score | 0.625384 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
NETR_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 5) | NC score | 0.417651 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
C163A_MOUSE
|
||||||
| θ value | 2.05525e-28 (rank : 6) | NC score | 0.850051 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
NETR_MOUSE
|
||||||
| θ value | 1.73987e-27 (rank : 7) | NC score | 0.407560 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
C163B_HUMAN
|
||||||
| θ value | 2.96777e-27 (rank : 8) | NC score | 0.845448 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
MSRE_MOUSE
|
||||||
| θ value | 1.92365e-26 (rank : 9) | NC score | 0.682843 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
LOXL2_MOUSE
|
||||||
| θ value | 1.24688e-25 (rank : 10) | NC score | 0.787721 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL4_HUMAN
|
||||||
| θ value | 6.18819e-25 (rank : 11) | NC score | 0.786472 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_HUMAN
|
||||||
| θ value | 1.05554e-24 (rank : 12) | NC score | 0.784644 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL4_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 13) | NC score | 0.785859 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
MSRE_HUMAN
|
||||||
| θ value | 2.59989e-23 (rank : 14) | NC score | 0.621260 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
CD6_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 15) | NC score | 0.840434 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |
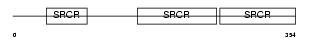
|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 3.75424e-22 (rank : 16) | NC score | 0.425536 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
MARCO_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 17) | NC score | 0.529654 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
LOXL3_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 18) | NC score | 0.778368 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL3_HUMAN
|
||||||
| θ value | 3.17815e-21 (rank : 19) | NC score | 0.777633 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
CD6_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 20) | NC score | 0.836317 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |
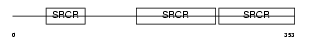
|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
CD5L_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 21) | NC score | 0.851096 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |
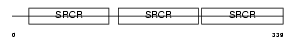
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
CD5L_MOUSE
|
||||||
| θ value | 8.95645e-16 (rank : 22) | NC score | 0.848219 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |
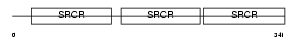
|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
KLHL5_HUMAN
|
||||||
| θ value | 3.41135e-07 (rank : 23) | NC score | 0.128964 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PQ7, Q86XW0, Q9NUK3, Q9NV27, Q9NVA9, Q9Y2X2 | Gene names | KLHL5 | |||
|
Domain Architecture |
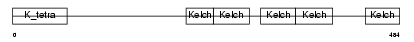
|
|||||
| Description | Kelch-like protein 5. | |||||
|
BTBD2_HUMAN
|
||||||
| θ value | 0.000602161 (rank : 24) | NC score | 0.143492 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |
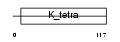
|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
KLH13_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 25) | NC score | 0.139951 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9P2N7, Q6P2E3, Q96QI7, Q9UDN9 | Gene names | KLHL13, BKLHD2, KIAA1309 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 13 (BTB and kelch domain-containing protein 2). | |||||
|
KLH13_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 26) | NC score | 0.140558 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TF4, Q9DBY7 | Gene names | Klhl13, Bklhd2, Kiaa1309 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 13 (BTB and kelch domain-containing protein 2). | |||||
|
KLHL9_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 27) | NC score | 0.137814 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6ZPT1 | Gene names | Klhl9, Kiaa1354 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 9. | |||||
|
KLHL9_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 28) | NC score | 0.137487 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9P2J3, Q8TCQ2 | Gene names | KLHL9, KIAA1354 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 9. | |||||
|
RCBT1_HUMAN
|
||||||
| θ value | 0.00298849 (rank : 29) | NC score | 0.112961 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
CFAI_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 30) | NC score | 0.118128 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
RCBT1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 31) | NC score | 0.111765 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
BTBD1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 32) | NC score | 0.136468 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H0C5, Q9BX71, Q9NWN4 | Gene names | BTBD1, C15orf1 | |||
|
Domain Architecture |
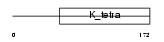
|
|||||
| Description | BTB/POZ domain-containing protein 1 (Hepatitis C virus NS5A- transactivated protein 8) (NS5ATP8). | |||||
|
BTBD1_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 33) | NC score | 0.136947 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58544, Q8BTZ0, Q8K0J0 | Gene names | Btbd1, Gsrp | |||
|
Domain Architecture |
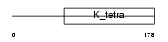
|
|||||
| Description | BTB/POZ domain-containing protein 1 (Glucose signal repressing protein). | |||||
|
KLH26_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 34) | NC score | 0.132232 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGY4, Q8CIG6, Q8R153 | Gene names | Klhl26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 26. | |||||
|
KLHL6_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 35) | NC score | 0.118200 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6V595 | Gene names | Klhl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 6. | |||||
|
KBTB3_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 36) | NC score | 0.128915 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BHI4, Q8BNJ8 | Gene names | Kbtbd3, Bklhd3 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 3 (BTB and kelch domain-containing protein 3). | |||||
|
KBTB3_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 37) | NC score | 0.128267 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NAB2, Q86X38, Q96NK5 | Gene names | KBTBD3, BKLHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 3 (BTB and kelch domain-containing protein 3). | |||||
|
KEAP1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 38) | NC score | 0.118590 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14145, Q6LEP0 | Gene names | KEAP1, KIAA0132, KLHL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like ECH-associated protein 1 (Cytosolic inhibitor of Nrf2) (Kelch-like protein 19). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 39) | NC score | 0.077199 (rank : 115) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
RCBT2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 40) | NC score | 0.103763 (rank : 86) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
BTBD9_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 41) | NC score | 0.131285 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96Q07, Q96M00 | Gene names | BTBD9, KIAA1880 | |||
|
Domain Architecture |
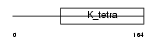
|
|||||
| Description | BTB/POZ domain-containing protein 9. | |||||
|
KEAP1_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 42) | NC score | 0.118603 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z2X8 | Gene names | Keap1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like ECH-associated protein 1 (Cytosolic inhibitor of Nrf2). | |||||
|
KLH26_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 43) | NC score | 0.128581 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q53HC5, Q8TAP0, Q9NUX3 | Gene names | KLHL26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 26. | |||||
|
RCBT2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 44) | NC score | 0.104858 (rank : 81) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
CD5_MOUSE
|
||||||
| θ value | 0.125558 (rank : 45) | NC score | 0.458510 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |
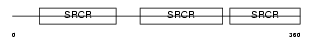
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
KLH20_HUMAN
|
||||||
| θ value | 0.125558 (rank : 46) | NC score | 0.115730 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2M5, Q5TZF2, Q9H457 | Gene names | KLHL20, KLEIP | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein) (Kelch- like protein X). | |||||
|
KLH20_MOUSE
|
||||||
| θ value | 0.125558 (rank : 47) | NC score | 0.115728 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VCK5, Q8BWA2 | Gene names | Klhl20, Kleip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein). | |||||
|
KLH22_HUMAN
|
||||||
| θ value | 0.125558 (rank : 48) | NC score | 0.129474 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q53GT1, Q96B68, Q96KC6 | Gene names | KLHL22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 22. | |||||
|
BTBD9_MOUSE
|
||||||
| θ value | 0.163984 (rank : 49) | NC score | 0.130046 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C726 | Gene names | Btbd9 | |||
|
Domain Architecture |
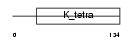
|
|||||
| Description | BTB/POZ domain-containing protein 9. | |||||
|
CFAI_MOUSE
|
||||||
| θ value | 0.163984 (rank : 50) | NC score | 0.114095 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
KLH14_HUMAN
|
||||||
| θ value | 0.279714 (rank : 51) | NC score | 0.120712 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
KLH22_MOUSE
|
||||||
| θ value | 0.62314 (rank : 52) | NC score | 0.127551 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99JN2, Q8BT13 | Gene names | Klhl22, Kelchl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 22. | |||||
|
CD5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 53) | NC score | 0.422147 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |
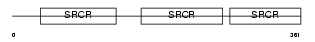
|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
IPP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 54) | NC score | 0.109271 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y573 | Gene names | IPP | |||
|
Domain Architecture |

|
|||||
| Description | Actin-binding protein IPP (MIPP protein). | |||||
|
KHL15_HUMAN
|
||||||
| θ value | 1.06291 (rank : 55) | NC score | 0.120633 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96M94, Q32MN3, Q8NDA3, Q96BM6, Q9C0I6 | Gene names | KLHL15, KIAA1677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 15. | |||||
|
KLH11_HUMAN
|
||||||
| θ value | 1.06291 (rank : 56) | NC score | 0.115187 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
ENTK_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.099502 (rank : 98) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
BKLH5_HUMAN
|
||||||
| θ value | 1.81305 (rank : 58) | NC score | 0.118711 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96NJ5, Q96PY7 | Gene names | BKLHD5, KIAA1900 | |||
|
Domain Architecture |

|
|||||
| Description | BTB and kelch domain-containing protein 5. | |||||
|
KLH11_MOUSE
|
||||||
| θ value | 1.81305 (rank : 59) | NC score | 0.114683 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CE33, Q3TDZ6, Q91VP6 | Gene names | Klhl11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
CNOT6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.014667 (rank : 177) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3P5, Q5NCL3, Q80V15 | Gene names | Cnot6, Ccr4, Kiaa1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.010532 (rank : 180) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
CNOT6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.014185 (rank : 178) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULM6 | Gene names | CNOT6, CCR4, KIAA1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
ENTK_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.087304 (rank : 109) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
RPTOR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.018872 (rank : 176) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N122, Q8N4V9, Q8TB32, Q9P2P3 | Gene names | RAPTOR, KIAA1303 | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
RPTOR_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.019057 (rank : 175) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4Q0, Q8C9W9, Q8CBY4, Q8CDY8, Q9D4H3 | Gene names | Raptor | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
TMPS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.079264 (rank : 114) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |
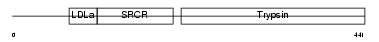
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
CTF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.012555 (rank : 179) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60753 | Gene names | Ctf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cardiotrophin-1 (CT-1). | |||||
|
ABTB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.075198 (rank : 122) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q969K4, Q6ZNU9, Q71MF1, Q96S62, Q96S63 | Gene names | ABTB1, BPOZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1 (Elongation factor 1A-binding protein). | |||||
|
ABTB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.069722 (rank : 129) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99LJ2, Q8C2K7, Q91XU4 | Gene names | Abtb1, Bpoz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1. | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.056386 (rank : 145) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ABTB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.054296 (rank : 150) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
BACH1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.057432 (rank : 140) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.057425 (rank : 141) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
BACH2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.059163 (rank : 136) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.058699 (rank : 137) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BTBD3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.109638 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2F9 | Gene names | BTBD3, KIAA0952 | |||
|
Domain Architecture |
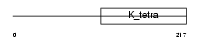
|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
BTBD3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.110005 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58545 | Gene names | Btbd3 | |||
|
Domain Architecture |
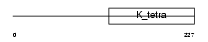
|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
BTBD5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.107976 (rank : 74) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NXS3 | Gene names | BTBD5 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 5. | |||||
|
BTBD5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.108470 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CR40, Q9CYV0 | Gene names | Btbd5 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 5. | |||||
|
BTBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.103613 (rank : 87) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96KE9, Q8IVQ7, Q9BR94 | Gene names | BTBD6, BDPL | |||
|
Domain Architecture |
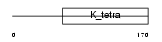
|
|||||
| Description | BTB/POZ domain-containing protein 6 (Lens BTB domain protein). | |||||
|
BTBD6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.104115 (rank : 84) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K2J9 | Gene names | Btbd6 | |||
|
Domain Architecture |
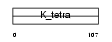
|
|||||
| Description | BTB/POZ domain-containing protein 6. | |||||
|
BTBD8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.085762 (rank : 111) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XKL5, Q6V9S5 | Gene names | BTBD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 8. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050377 (rank : 172) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.050757 (rank : 171) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.052989 (rank : 160) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CALI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.095286 (rank : 104) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13939, Q9BXG7 | Gene names | CCIN | |||
|
Domain Architecture |

|
|||||
| Description | Calicin. | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.050758 (rank : 170) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.051685 (rank : 168) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CO6A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.050274 (rank : 174) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |
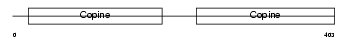
|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
CO9A1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.053168 (rank : 157) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.052614 (rank : 162) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.053041 (rank : 158) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.053779 (rank : 153) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.052954 (rank : 161) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
COJA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056791 (rank : 143) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
COLQ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.056842 (rank : 142) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y215, Q6DK18, Q6YH18, Q6YH19, Q6YH20, Q6YH21, Q9NP18, Q9NP19, Q9NP20, Q9NP21, Q9NP22, Q9NP23, Q9NP24, Q9UP88 | Gene names | COLQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acetylcholinesterase collagenic tail peptide precursor (AChE Q subunit) (Acetylcholinesterase-associated collagen). | |||||
|
CONA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.054524 (rank : 149) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CONA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.053912 (rank : 152) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4G2, Q5SUQ0 | Gene names | Col23a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CORIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.053315 (rank : 155) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CS1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.052529 (rank : 163) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.053232 (rank : 156) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CUZD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.094628 (rank : 105) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
CUZD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.097723 (rank : 102) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
ENC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.101791 (rank : 92) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14682, O75464, Q9UPG9 | Gene names | ENC1, NRPB, PIG10 | |||
|
Domain Architecture |

|
|||||
| Description | Ectoderm-neural cortex 1 protein (ENC-1) (p53-induced protein 10) (Nuclear matrix protein NRP/B). | |||||
|
ENC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.101746 (rank : 93) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35709 | Gene names | Enc1, Enc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectoderm-neural cortex 1 protein (ENC-1). | |||||
|
GAN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.109358 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2C0 | Gene names | GAN, GAN1, KLHL16 | |||
|
Domain Architecture |

|
|||||
| Description | Gigaxonin (Kelch-like protein 16). | |||||
|
GMCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.064190 (rank : 134) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96IK5, Q9H826, Q9H8V7, Q9H927 | Gene names | GMCL1, GCL | |||
|
Domain Architecture |
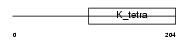
|
|||||
| Description | Germ cell-less protein-like 1. | |||||
|
GMCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.065329 (rank : 132) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q920G9, Q9DBW5, Q9QUP6 | Gene names | Gmcl1, Gcl, Gcl1 | |||
|
Domain Architecture |
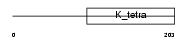
|
|||||
| Description | Germ cell-less protein-like 1 (mGcl-1) (DP-interacting protein) (DIP). | |||||
|
GMCLL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.052356 (rank : 164) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NEA9, Q8TC88, Q8TC89 | Gene names | GMCL1L, GCL | |||
|
Domain Architecture |
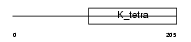
|
|||||
| Description | Germ cell-less protein-like 1-like. | |||||
|
IPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.108064 (rank : 73) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28575 | Gene names | Ipp, Mipp | |||
|
Domain Architecture |

|
|||||
| Description | Actin-binding protein IPP (MIPP protein) (Murine IAP-promoted placenta-expressed protein). | |||||
|
KBTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.108544 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IY47, Q86Y62, Q9P239, Q9UFM7 | Gene names | KBTBD2, BKLHD1, KIAA1489 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 2 (BTB and kelch domain-containing protein 1). | |||||
|
KBTB4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.099697 (rank : 97) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |
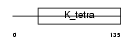
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
KBTB4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.099838 (rank : 96) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R179, Q9CQX1 | Gene names | Kbtbd4, Bklhd4 | |||
|
Domain Architecture |
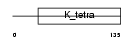
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
KBTB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.099250 (rank : 99) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86V97, Q8N8L0, Q8NDM5, Q96MP6 | Gene names | KBTBD6 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 6. | |||||
|
KBTB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.101950 (rank : 91) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WVZ9, Q8NB99, Q9H0I6 | Gene names | KBTBD7 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 7. | |||||
|
KBTB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.098672 (rank : 101) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96CT2, Q8N388, Q96BF0, Q96PW7 | Gene names | KBTBD9, KIAA1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
KBTB9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.098768 (rank : 100) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80T74 | Gene names | Kbtbd9, Kiaa1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
KBTBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.109857 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60662 | Gene names | KBTBD10, KRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 10 (Kelch-related protein 1) (Kel-like protein 23) (Sarcosin). | |||||
|
KBTBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.092014 (rank : 107) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94819 | Gene names | KBTBD11, KIAA0711, KLHDC7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch repeat and BTB domain-containing protein 11 (Kelch domain- containing protein 7B). | |||||
|
KBTBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.091247 (rank : 108) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BNW9 | Gene names | Kbtbd11, Kiaa0711 | |||
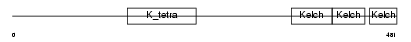
|
Domain Architecture |
|
|||||
| Description | Kelch repeat and BTB domain-containing protein 11. | |||||
|
KLD7B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.086127 (rank : 110) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96G42 | Gene names | KLHDC7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 7B. | |||||
|
KLD8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.073121 (rank : 124) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IYD2, Q9NVG5 | Gene names | KLHDC8A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8A. | |||||
|
KLD8A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.076424 (rank : 118) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91XA8 | Gene names | Klhdc8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8A. | |||||
|
KLD8B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.072287 (rank : 126) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IXV7 | Gene names | KLHDC8B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8B. | |||||
|
KLD8B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.072320 (rank : 125) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D2D9 | Gene names | Klhdc8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8B. | |||||
|
KLDC5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.096331 (rank : 103) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2K6, Q2VPK1, Q8N334 | Gene names | KLHDC5, KIAA1340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 5. | |||||
|
KLDC5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.094018 (rank : 106) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BFQ9, Q6ZPT4 | Gene names | Klhdc5, Kiaa1340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 5. | |||||
|
KLH10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.108359 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6JEL2, Q6NW28, Q96MC0 | Gene names | KLHL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 10. | |||||
|
KLH10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.108586 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D5V2, Q8BVM3, Q9DA07 | Gene names | Klhl10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 10. | |||||
|
KLH12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.101173 (rank : 95) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q53G59, Q9HBX5 | Gene names | KLHL12, C3IP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 12 (CUL3-interacting protein 1). | |||||
|
KLH12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.101318 (rank : 94) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BZM0, Q8K225 | Gene names | Klhl12, C3ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 12 (CUL3-interacting protein 1). | |||||
|
KLH17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.101994 (rank : 90) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6TDP4 | Gene names | KLHL17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 17 (Actinfilin). | |||||
|
KLH17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.102080 (rank : 89) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6TDP3 | Gene names | Klhl17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 17 (Actinfilin). | |||||
|
KLH18_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.105884 (rank : 80) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94889, Q8N125 | Gene names | KLHL18, KIAA0795 | |||
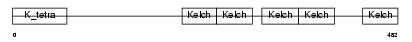
|
Domain Architecture |
|
|||||
| Description | Kelch-like protein 18. | |||||
|
KLH21_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.102741 (rank : 88) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UJP4, O75057 | Gene names | KLHL21, KIAA0469 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 21. | |||||
|
KLH23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.106140 (rank : 79) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NBE8, Q8N9B9, Q96FT8 | Gene names | KLHL23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 23. | |||||
|
KLH23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.106480 (rank : 78) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6GQU2, Q8C840 | Gene names | Klhl23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 23. | |||||
|
KLHL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.106853 (rank : 77) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NR64, Q5VZ64, Q9H4X4, Q9NR65, Q9P238 | Gene names | KLHL1, KIAA1490 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 1. | |||||
|
KLHL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.107019 (rank : 76) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JI74 | Gene names | Klhl1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 1. | |||||
|
KLHL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.104023 (rank : 85) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95198, Q8N484, Q8TBH5 | Gene names | KLHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 2 (Actin-binding protein Mayven). | |||||
|
KLHL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.104377 (rank : 82) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8JZP3, Q8CCU0, Q8R3U4 | Gene names | Klhl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 2. | |||||
|
KLHL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.104158 (rank : 83) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UH77, Q9UH75, Q9UH76, Q9ULU0, Q9Y6V6 | Gene names | KLHL3, KIAA1129 | |||
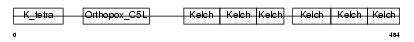
|
Domain Architecture |
|
|||||
| Description | Kelch-like protein 3. | |||||
|
KLHL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.108347 (rank : 72) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9C0H6, Q9Y3J5 | Gene names | KLHL4, KIAA1687 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 4. | |||||
|
KLHL6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.110226 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ60, Q8N5I1, Q8N892 | Gene names | KLHL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 6. | |||||
|
KLHL7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.110903 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IXQ5, Q9BQF8 | Gene names | KLHL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 7. | |||||
|
KLHL7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.110574 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BUL5, Q9CZP4 | Gene names | Klhl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 7. | |||||
|
KLHL8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.107942 (rank : 75) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P2G9 | Gene names | KLHL8, KIAA1378 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 8. | |||||
|
KLHL8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.108351 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59280 | Gene names | Klhl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 8. | |||||
|
LOXL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.275250 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.274284 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
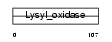
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
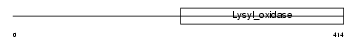
LYOX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.289364 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
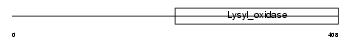
LYOX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.287175 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |
|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LZTR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.076249 (rank : 119) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N653, Q14776 | Gene names | LZTR1, TCFL2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
LZTR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.076177 (rank : 120) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQ33 | Gene names | Lztr1, Tcfl2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.053558 (rank : 154) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.053012 (rank : 159) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.052342 (rank : 165) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
PCOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.051202 (rank : 169) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
PCOC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.051780 (rank : 167) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
RHBT3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.076524 (rank : 117) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94955, Q8IW06 | Gene names | RHOBTB3, KIAA0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
RHBT3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.076686 (rank : 116) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CTN4, Q80X55, Q9CVT0 | Gene names | Rhobtb3, Kiaa0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
SCAR3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.063154 (rank : 135) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6AZY7, Q9UM15, Q9UM16 | Gene names | SCARA3, CSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3 (Cellular stress response gene protein). | |||||
|
SCAR3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.064426 (rank : 133) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C850, Q8C6N1 | Gene names | Scara3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3. | |||||
|
SPOP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.080244 (rank : 112) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43791 | Gene names | SPOP | |||
|
Domain Architecture |
|
|||||
| Description | Speckle-type POZ protein. | |||||
|
SPOP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.080244 (rank : 113) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZWS8 | Gene names | Spop, Pcif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Speckle-type POZ protein (PDX-1 C-terminal-interacting factor 1) (PCIF1). | |||||
|
ST14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.056469 (rank : 144) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ST14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.058476 (rank : 138) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TDPZ1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.070463 (rank : 127) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q717B3, Q71G58, Q8BZV7, Q8VI39 | Gene names | Tdpoz1, 2cpoz56, Mapp2, Spopl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 1 (MAPP family protein 2). | |||||
|
TDPZ2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.070404 (rank : 128) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q717B2 | Gene names | Tdpoz2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 2. | |||||
|
TDPZ3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.065927 (rank : 130) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q717B4 | Gene names | Tdpoz3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 3. | |||||
|
TDPZ4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.073560 (rank : 123) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6YCH2 | Gene names | Tdpoz4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 4. | |||||
|
TDPZ5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.065922 (rank : 131) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6YCH1 | Gene names | Tdpoz5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 5. | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.054180 (rank : 151) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.075301 (rank : 121) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.058402 (rank : 139) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |
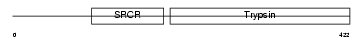
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
TMPS5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.054537 (rank : 148) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |
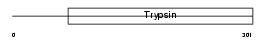
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.051952 (rank : 166) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.055124 (rank : 147) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ZP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.055329 (rank : 146) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
ZP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050363 (rank : 173) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
LG3BP_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q08380 | Gene names | LGALS3BP, M2BP | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3-binding protein precursor (Lectin galactoside-binding soluble 3-binding protein) (Mac-2-binding protein) (Mac-2 BP) (MAC2BP) (Tumor-associated antigen 90K). | |||||
|
C163A_HUMAN
|
||||||
| NC score | 0.854556 (rank : 2) | θ value | 1.52067e-31 (rank : 2) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86VB7, Q07898, Q07899, Q07900, Q07901, Q2VLH7 | Gene names | CD163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) (Hemoglobin scavenger receptor) [Contains: Soluble CD163 (sCD163)]. | |||||
|
CD5L_HUMAN
|
||||||
| NC score | 0.851096 (rank : 3) | θ value | 2.35696e-16 (rank : 21) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O43866, Q6UX63 | Gene names | CD5L | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (IgM-associated peptide). | |||||
|
C163A_MOUSE
|
||||||
| NC score | 0.850051 (rank : 4) | θ value | 2.05525e-28 (rank : 6) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q2VLH6, Q2VLH5, Q99MX8 | Gene names | Cd163, M130 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M130 precursor (CD163 antigen) [Contains: Soluble CD163 (sCD163)]. | |||||
|
CD5L_MOUSE
|
||||||
| NC score | 0.848219 (rank : 5) | θ value | 8.95645e-16 (rank : 22) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9QWK4, O35300, O35301, Q505P6, Q91W05 | Gene names | Cd5l, Aim, Api6 | |||
|
Domain Architecture |

|
|||||
| Description | CD5 antigen-like precursor (SP-alpha) (CT-2) (Apoptosis inhibitory 6) (Apoptosis inhibitor expressed by macrophages). | |||||
|
C163B_HUMAN
|
||||||
| NC score | 0.845448 (rank : 6) | θ value | 2.96777e-27 (rank : 8) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NR16, Q6UWC2 | Gene names | CD163L1, CD163B, M160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor cysteine-rich type 1 protein M160 precursor (CD163 antigen-like 1) (CD163b antigen). | |||||
|
CD6_HUMAN
|
||||||
| NC score | 0.840434 (rank : 7) | θ value | 3.75424e-22 (rank : 15) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P30203, Q9UMF2, Q9Y4K7, Q9Y4K8, Q9Y4K9, Q9Y4L0 | Gene names | CD6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor (T12) (TP120). | |||||
|
CD6_MOUSE
|
||||||
| NC score | 0.836317 (rank : 8) | θ value | 2.06002e-20 (rank : 20) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61003, Q60679, Q61004 | Gene names | Cd6 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell differentiation antigen CD6 precursor. | |||||
|
LOXL2_MOUSE
|
||||||
| NC score | 0.787721 (rank : 9) | θ value | 1.24688e-25 (rank : 10) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P58022, Q8BRE6, Q8BS86, Q8C4K0, Q9JJ39 | Gene names | Loxl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2). | |||||
|
LOXL4_HUMAN
|
||||||
| NC score | 0.786472 (rank : 10) | θ value | 6.18819e-25 (rank : 11) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96JB6, Q96DY1, Q96PC0, Q9H6T5 | Gene names | LOXL4, LOXC | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL4_MOUSE
|
||||||
| NC score | 0.785859 (rank : 11) | θ value | 3.07116e-24 (rank : 13) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q924C6 | Gene names | Loxl4, Loxc | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 4 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 4) (Lysyl oxidase-related protein C). | |||||
|
LOXL2_HUMAN
|
||||||
| NC score | 0.784644 (rank : 12) | θ value | 1.05554e-24 (rank : 12) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y4K0, Q9BW70, Q9Y5Y8 | Gene names | LOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 2 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 2) (Lysyl oxidase-related protein 2) (Lysyl oxidase-related protein WS9-14). | |||||
|
LOXL3_MOUSE
|
||||||
| NC score | 0.778368 (rank : 13) | θ value | 1.42661e-21 (rank : 18) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Z175, Q9JJ39 | Gene names | Loxl3, Lor2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3) (Lysyl oxidase-related protein 2). | |||||
|
LOXL3_HUMAN
|
||||||
| NC score | 0.777633 (rank : 14) | θ value | 3.17815e-21 (rank : 19) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P58215, Q96RS1 | Gene names | LOXL3, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 3 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 3). | |||||
|
DMBT1_HUMAN
|
||||||
| NC score | 0.685159 (rank : 15) | θ value | 2.59387e-31 (rank : 3) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9UGM3, Q59EX0, Q5JR26, Q6MZN4, Q96DU4, Q9UGM2, Q9UJ57, Q9UKJ4, Q9Y211, Q9Y4V9 | Gene names | DMBT1, GP340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (Glycoprotein 340) (Gp-340) (Surfactant pulmonary-associated D-binding protein). | |||||
|
MSRE_MOUSE
|
||||||
| NC score | 0.682843 (rank : 16) | θ value | 1.92365e-26 (rank : 9) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P30204, Q923G0, Q9QZ56 | Gene names | Msr1, Scvr | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor type A) (SR-A). | |||||
|
DMBT1_MOUSE
|
||||||
| NC score | 0.625384 (rank : 17) | θ value | 9.8567e-31 (rank : 4) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q60997, Q80YC6, Q9JMJ9 | Gene names | Dmbt1, Crpd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Deleted in malignant brain tumors 1 protein precursor (CRP-ductin) (Vomeroglandin). | |||||
|
MSRE_HUMAN
|
||||||
| NC score | 0.621260 (rank : 18) | θ value | 2.59989e-23 (rank : 14) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P21757, P21759, Q45F10 | Gene names | MSR1, SCARA1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage scavenger receptor types I and II (Macrophage acetylated LDL receptor I and II) (Scavenger receptor class A member 1) (CD204 antigen). | |||||
|
MARCO_HUMAN
|
||||||
| NC score | 0.529654 (rank : 19) | θ value | 1.09232e-21 (rank : 17) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UEW3, Q9Y5S3 | Gene names | MARCO, SCARA2 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure) (Scavenger receptor class A member 2). | |||||
|
CD5_MOUSE
|
||||||
| NC score | 0.458510 (rank : 20) | θ value | 0.125558 (rank : 45) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13379 | Gene names | Cd5, Ly-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen Ly-1) (Lyt-1). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | 0.425536 (rank : 21) | θ value | 3.75424e-22 (rank : 16) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
CD5_HUMAN
|
||||||
| NC score | 0.422147 (rank : 22) | θ value | 0.813845 (rank : 53) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P06127 | Gene names | CD5, LEU1 | |||
|
Domain Architecture |

|
|||||
| Description | T-cell surface glycoprotein CD5 precursor (Lymphocyte antigen T1/Leu- 1). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.417651 (rank : 23) | θ value | 5.40856e-29 (rank : 5) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.407560 (rank : 24) | θ value | 1.73987e-27 (rank : 7) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
LYOX_HUMAN
|
||||||
| NC score | 0.289364 (rank : 25) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28300, Q5FWF0 | Gene names | LOX | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase). | |||||
|
LYOX_MOUSE
|
||||||
| NC score | 0.287175 (rank : 26) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28301 | Gene names | Lox, Rrg | |||
|
Domain Architecture |

|
|||||
| Description | Protein-lysine 6-oxidase precursor (EC 1.4.3.13) (Lysyl oxidase) (RAS excision protein). | |||||
|
LOXL1_HUMAN
|
||||||
| NC score | 0.275250 (rank : 27) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
LOXL1_MOUSE
|
||||||
| NC score | 0.274284 (rank : 28) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97873, Q91ZY4, Q99KX3 | Gene names | Loxl1, Lox2, Loxl | |||
|
Domain Architecture |
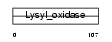
|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (Lysyl oxidase 2). | |||||
|
BTBD2_HUMAN
|
||||||
| NC score | 0.143492 (rank : 29) | θ value | 0.000602161 (rank : 24) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9BX70, O60418, O75248, Q7Z5W0, Q96SX8, Q9NPS1, Q9NX81 | Gene names | BTBD2 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 2. | |||||
|
KLH13_MOUSE
|
||||||
| NC score | 0.140558 (rank : 30) | θ value | 0.000786445 (rank : 26) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q80TF4, Q9DBY7 | Gene names | Klhl13, Bklhd2, Kiaa1309 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 13 (BTB and kelch domain-containing protein 2). | |||||
|
KLH13_HUMAN
|
||||||
| NC score | 0.139951 (rank : 31) | θ value | 0.000786445 (rank : 25) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9P2N7, Q6P2E3, Q96QI7, Q9UDN9 | Gene names | KLHL13, BKLHD2, KIAA1309 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 13 (BTB and kelch domain-containing protein 2). | |||||
|
KLHL9_MOUSE
|
||||||
| NC score | 0.137814 (rank : 32) | θ value | 0.00134147 (rank : 27) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6ZPT1 | Gene names | Klhl9, Kiaa1354 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 9. | |||||
|
KLHL9_HUMAN
|
||||||
| NC score | 0.137487 (rank : 33) | θ value | 0.00298849 (rank : 28) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9P2J3, Q8TCQ2 | Gene names | KLHL9, KIAA1354 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 9. | |||||
|
BTBD1_MOUSE
|
||||||
| NC score | 0.136947 (rank : 34) | θ value | 0.0113563 (rank : 33) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58544, Q8BTZ0, Q8K0J0 | Gene names | Btbd1, Gsrp | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 1 (Glucose signal repressing protein). | |||||
|
BTBD1_HUMAN
|
||||||
| NC score | 0.136468 (rank : 35) | θ value | 0.00869519 (rank : 32) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H0C5, Q9BX71, Q9NWN4 | Gene names | BTBD1, C15orf1 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 1 (Hepatitis C virus NS5A- transactivated protein 8) (NS5ATP8). | |||||
|
KLH26_MOUSE
|
||||||
| NC score | 0.132232 (rank : 36) | θ value | 0.0193708 (rank : 34) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BGY4, Q8CIG6, Q8R153 | Gene names | Klhl26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 26. | |||||
|
BTBD9_HUMAN
|
||||||
| NC score | 0.131285 (rank : 37) | θ value | 0.0736092 (rank : 41) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96Q07, Q96M00 | Gene names | BTBD9, KIAA1880 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 9. | |||||
|
BTBD9_MOUSE
|
||||||
| NC score | 0.130046 (rank : 38) | θ value | 0.163984 (rank : 49) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8C726 | Gene names | Btbd9 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 9. | |||||
|
KLH22_HUMAN
|
||||||
| NC score | 0.129474 (rank : 39) | θ value | 0.125558 (rank : 48) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q53GT1, Q96B68, Q96KC6 | Gene names | KLHL22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 22. | |||||
|
KLHL5_HUMAN
|
||||||
| NC score | 0.128964 (rank : 40) | θ value | 3.41135e-07 (rank : 23) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96PQ7, Q86XW0, Q9NUK3, Q9NV27, Q9NVA9, Q9Y2X2 | Gene names | KLHL5 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 5. | |||||
|
KBTB3_MOUSE
|
||||||
| NC score | 0.128915 (rank : 41) | θ value | 0.0252991 (rank : 36) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8BHI4, Q8BNJ8 | Gene names | Kbtbd3, Bklhd3 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 3 (BTB and kelch domain-containing protein 3). | |||||
|
KLH26_HUMAN
|
||||||
| NC score | 0.128581 (rank : 42) | θ value | 0.0736092 (rank : 43) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q53HC5, Q8TAP0, Q9NUX3 | Gene names | KLHL26 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 26. | |||||
|
KBTB3_HUMAN
|
||||||
| NC score | 0.128267 (rank : 43) | θ value | 0.0330416 (rank : 37) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NAB2, Q86X38, Q96NK5 | Gene names | KBTBD3, BKLHD3 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 3 (BTB and kelch domain-containing protein 3). | |||||
|
KLH22_MOUSE
|
||||||
| NC score | 0.127551 (rank : 44) | θ value | 0.62314 (rank : 52) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q99JN2, Q8BT13 | Gene names | Klhl22, Kelchl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 22. | |||||
|
KLH14_HUMAN
|
||||||
| NC score | 0.120712 (rank : 45) | θ value | 0.279714 (rank : 51) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9P2G3 | Gene names | KLHL14, KIAA1384 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 14. | |||||
|
KHL15_HUMAN
|
||||||
| NC score | 0.120633 (rank : 46) | θ value | 1.06291 (rank : 55) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q96M94, Q32MN3, Q8NDA3, Q96BM6, Q9C0I6 | Gene names | KLHL15, KIAA1677 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 15. | |||||
|
BKLH5_HUMAN
|
||||||
| NC score | 0.118711 (rank : 47) | θ value | 1.81305 (rank : 58) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96NJ5, Q96PY7 | Gene names | BKLHD5, KIAA1900 | |||
|
Domain Architecture |

|
|||||
| Description | BTB and kelch domain-containing protein 5. | |||||
|
KEAP1_MOUSE
|
||||||
| NC score | 0.118603 (rank : 48) | θ value | 0.0736092 (rank : 42) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z2X8 | Gene names | Keap1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like ECH-associated protein 1 (Cytosolic inhibitor of Nrf2). | |||||
|
KEAP1_HUMAN
|
||||||
| NC score | 0.118590 (rank : 49) | θ value | 0.0431538 (rank : 38) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q14145, Q6LEP0 | Gene names | KEAP1, KIAA0132, KLHL19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like ECH-associated protein 1 (Cytosolic inhibitor of Nrf2) (Kelch-like protein 19). | |||||
|
KLHL6_MOUSE
|
||||||
| NC score | 0.118200 (rank : 50) | θ value | 0.0193708 (rank : 35) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6V595 | Gene names | Klhl6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 6. | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.118128 (rank : 51) | θ value | 0.00509761 (rank : 30) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
KLH20_HUMAN
|
||||||
| NC score | 0.115730 (rank : 52) | θ value | 0.125558 (rank : 46) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y2M5, Q5TZF2, Q9H457 | Gene names | KLHL20, KLEIP | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein) (Kelch- like protein X). | |||||
|
KLH20_MOUSE
|
||||||
| NC score | 0.115728 (rank : 53) | θ value | 0.125558 (rank : 47) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VCK5, Q8BWA2 | Gene names | Klhl20, Kleip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 20 (Kelch-like ECT2-interacting protein). | |||||
|
KLH11_HUMAN
|
||||||
| NC score | 0.115187 (rank : 54) | θ value | 1.06291 (rank : 56) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NVR0 | Gene names | KLHL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
KLH11_MOUSE
|
||||||
| NC score | 0.114683 (rank : 55) | θ value | 1.81305 (rank : 59) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CE33, Q3TDZ6, Q91VP6 | Gene names | Klhl11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 11 precursor. | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.114095 (rank : 56) | θ value | 0.163984 (rank : 50) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
RCBT1_HUMAN
|
||||||
| NC score | 0.112961 (rank : 57) | θ value | 0.00298849 (rank : 29) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8NDN9, Q969U9 | Gene names | RCBTB1, CLLD7, E4.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1) (Chronic lymphocytic leukemia deletion region gene 7 protein) (CLL deletion region gene 7 protein). | |||||
|
RCBT1_MOUSE
|
||||||
| NC score | 0.111765 (rank : 58) | θ value | 0.00509761 (rank : 31) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6NXM2, Q8BTZ6, Q8BZV0 | Gene names | Rcbtb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 1 (Regulator of chromosome condensation and BTB domain-containing protein 1). | |||||
|
KLHL7_HUMAN
|
||||||
| NC score | 0.110903 (rank : 59) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IXQ5, Q9BQF8 | Gene names | KLHL7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 7. | |||||
|
KLHL7_MOUSE
|
||||||
| NC score | 0.110574 (rank : 60) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BUL5, Q9CZP4 | Gene names | Klhl7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 7. | |||||
|
KLHL6_HUMAN
|
||||||
| NC score | 0.110226 (rank : 61) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ60, Q8N5I1, Q8N892 | Gene names | KLHL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 6. | |||||
|
BTBD3_MOUSE
|
||||||
| NC score | 0.110005 (rank : 62) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P58545 | Gene names | Btbd3 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
KBTBA_HUMAN
|
||||||
| NC score | 0.109857 (rank : 63) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O60662 | Gene names | KBTBD10, KRP1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 10 (Kelch-related protein 1) (Kel-like protein 23) (Sarcosin). | |||||
|
BTBD3_HUMAN
|
||||||
| NC score | 0.109638 (rank : 64) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Y2F9 | Gene names | BTBD3, KIAA0952 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 3. | |||||
|
GAN_HUMAN
|
||||||
| NC score | 0.109358 (rank : 65) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2C0 | Gene names | GAN, GAN1, KLHL16 | |||
|
Domain Architecture |

|
|||||
| Description | Gigaxonin (Kelch-like protein 16). | |||||
|
IPP_HUMAN
|
||||||
| NC score | 0.109271 (rank : 66) | θ value | 1.06291 (rank : 54) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y573 | Gene names | IPP | |||
|
Domain Architecture |

|
|||||
| Description | Actin-binding protein IPP (MIPP protein). | |||||
|
KLH10_MOUSE
|
||||||
| NC score | 0.108586 (rank : 67) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9D5V2, Q8BVM3, Q9DA07 | Gene names | Klhl10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 10. | |||||
|
KBTB2_HUMAN
|
||||||
| NC score | 0.108544 (rank : 68) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8IY47, Q86Y62, Q9P239, Q9UFM7 | Gene names | KBTBD2, BKLHD1, KIAA1489 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 2 (BTB and kelch domain-containing protein 1). | |||||
|
BTBD5_MOUSE
|
||||||
| NC score | 0.108470 (rank : 69) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CR40, Q9CYV0 | Gene names | Btbd5 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 5. | |||||
|
KLH10_HUMAN
|
||||||
| NC score | 0.108359 (rank : 70) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6JEL2, Q6NW28, Q96MC0 | Gene names | KLHL10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 10. | |||||
|
KLHL8_MOUSE
|
||||||
| NC score | 0.108351 (rank : 71) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P59280 | Gene names | Klhl8 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 8. | |||||
|
KLHL4_HUMAN
|
||||||
| NC score | 0.108347 (rank : 72) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9C0H6, Q9Y3J5 | Gene names | KLHL4, KIAA1687 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 4. | |||||
|
IPP_MOUSE
|
||||||
| NC score | 0.108064 (rank : 73) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P28575 | Gene names | Ipp, Mipp | |||
|
Domain Architecture |

|
|||||
| Description | Actin-binding protein IPP (MIPP protein) (Murine IAP-promoted placenta-expressed protein). | |||||
|
BTBD5_HUMAN
|
||||||
| NC score | 0.107976 (rank : 74) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9NXS3 | Gene names | BTBD5 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 5. | |||||
|
KLHL8_HUMAN
|
||||||
| NC score | 0.107942 (rank : 75) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9P2G9 | Gene names | KLHL8, KIAA1378 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 8. | |||||
|
KLHL1_MOUSE
|
||||||
| NC score | 0.107019 (rank : 76) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9JI74 | Gene names | Klhl1 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 1. | |||||
|
KLHL1_HUMAN
|
||||||
| NC score | 0.106853 (rank : 77) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NR64, Q5VZ64, Q9H4X4, Q9NR65, Q9P238 | Gene names | KLHL1, KIAA1490 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 1. | |||||
|
KLH23_MOUSE
|
||||||
| NC score | 0.106480 (rank : 78) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6GQU2, Q8C840 | Gene names | Klhl23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 23. | |||||
|
KLH23_HUMAN
|
||||||
| NC score | 0.106140 (rank : 79) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NBE8, Q8N9B9, Q96FT8 | Gene names | KLHL23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 23. | |||||
|
KLH18_HUMAN
|
||||||
| NC score | 0.105884 (rank : 80) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94889, Q8N125 | Gene names | KLHL18, KIAA0795 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 18. | |||||
|
RCBT2_HUMAN
|
||||||
| NC score | 0.104858 (rank : 81) | θ value | 0.0961366 (rank : 44) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O95199 | Gene names | RCBTB2, CHC1L, RLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like) (CHC1-L) (RCC1-like G exchanging factor). | |||||
|
KLHL2_MOUSE
|
||||||
| NC score | 0.104377 (rank : 82) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8JZP3, Q8CCU0, Q8R3U4 | Gene names | Klhl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 2. | |||||
|
KLHL3_HUMAN
|
||||||
| NC score | 0.104158 (rank : 83) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UH77, Q9UH75, Q9UH76, Q9ULU0, Q9Y6V6 | Gene names | KLHL3, KIAA1129 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 3. | |||||
|
BTBD6_MOUSE
|
||||||
| NC score | 0.104115 (rank : 84) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K2J9 | Gene names | Btbd6 | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 6. | |||||
|
KLHL2_HUMAN
|
||||||
| NC score | 0.104023 (rank : 85) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O95198, Q8N484, Q8TBH5 | Gene names | KLHL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 2 (Actin-binding protein Mayven). | |||||
|
RCBT2_MOUSE
|
||||||
| NC score | 0.103763 (rank : 86) | θ value | 0.0563607 (rank : 40) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LJ7, Q3TUA3, Q8BMG2 | Gene names | Rcbtb2, Chc1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RCC1 and BTB domain-containing protein 2 (Regulator of chromosome condensation and BTB domain-containing protein 2) (Chromosome condensation 1-like). | |||||
|
BTBD6_HUMAN
|
||||||
| NC score | 0.103613 (rank : 87) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96KE9, Q8IVQ7, Q9BR94 | Gene names | BTBD6, BDPL | |||
|
Domain Architecture |

|
|||||
| Description | BTB/POZ domain-containing protein 6 (Lens BTB domain protein). | |||||
|
KLH21_HUMAN
|
||||||
| NC score | 0.102741 (rank : 88) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9UJP4, O75057 | Gene names | KLHL21, KIAA0469 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch-like protein 21. | |||||
|
KLH17_MOUSE
|
||||||
| NC score | 0.102080 (rank : 89) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6TDP3 | Gene names | Klhl17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 17 (Actinfilin). | |||||
|
KLH17_HUMAN
|
||||||
| NC score | 0.101994 (rank : 90) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q6TDP4 | Gene names | KLHL17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 17 (Actinfilin). | |||||
|
KBTB7_HUMAN
|
||||||
| NC score | 0.101950 (rank : 91) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WVZ9, Q8NB99, Q9H0I6 | Gene names | KBTBD7 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 7. | |||||
|
ENC1_HUMAN
|
||||||
| NC score | 0.101791 (rank : 92) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14682, O75464, Q9UPG9 | Gene names | ENC1, NRPB, PIG10 | |||
|
Domain Architecture |

|
|||||
| Description | Ectoderm-neural cortex 1 protein (ENC-1) (p53-induced protein 10) (Nuclear matrix protein NRP/B). | |||||
|
ENC1_MOUSE
|
||||||
| NC score | 0.101746 (rank : 93) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O35709 | Gene names | Enc1, Enc-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ectoderm-neural cortex 1 protein (ENC-1). | |||||
|
KLH12_MOUSE
|
||||||
| NC score | 0.101318 (rank : 94) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BZM0, Q8K225 | Gene names | Klhl12, C3ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 12 (CUL3-interacting protein 1). | |||||
|
KLH12_HUMAN
|
||||||
| NC score | 0.101173 (rank : 95) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q53G59, Q9HBX5 | Gene names | KLHL12, C3IP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch-like protein 12 (CUL3-interacting protein 1). | |||||
|
KBTB4_MOUSE
|
||||||
| NC score | 0.099838 (rank : 96) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8R179, Q9CQX1 | Gene names | Kbtbd4, Bklhd4 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
KBTB4_HUMAN
|
||||||
| NC score | 0.099697 (rank : 97) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NVX7, Q6IA85, Q9BUC3, Q9NV76 | Gene names | KBTBD4, BKLHD4 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 4 (BTB and kelch domain-containing protein 4). | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.099502 (rank : 98) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
KBTB6_HUMAN
|
||||||
| NC score | 0.099250 (rank : 99) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q86V97, Q8N8L0, Q8NDM5, Q96MP6 | Gene names | KBTBD6 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 6. | |||||
|
KBTB9_MOUSE
|
||||||
| NC score | 0.098768 (rank : 100) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q80T74 | Gene names | Kbtbd9, Kiaa1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
KBTB9_HUMAN
|
||||||
| NC score | 0.098672 (rank : 101) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96CT2, Q8N388, Q96BF0, Q96PW7 | Gene names | KBTBD9, KIAA1921 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 9. | |||||
|
CUZD1_MOUSE
|
||||||
| NC score | 0.097723 (rank : 102) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70412, Q9CTZ7 | Gene names | Cuzd1, Itmap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Integral membrane-associated protein 1). | |||||
|
KLDC5_HUMAN
|
||||||
| NC score | 0.096331 (rank : 103) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9P2K6, Q2VPK1, Q8N334 | Gene names | KLHDC5, KIAA1340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 5. | |||||
|
CALI_HUMAN
|
||||||
| NC score | 0.095286 (rank : 104) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q13939, Q9BXG7 | Gene names | CCIN | |||
|
Domain Architecture |

|
|||||
| Description | Calicin. | |||||
|
CUZD1_HUMAN
|
||||||
| NC score | 0.094628 (rank : 105) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UP6, Q7Z660, Q7Z661, Q86SG1, Q86UP5, Q9HAR7 | Gene names | CUZD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CUB and zona pellucida-like domain-containing protein 1 precursor (Transmembrane protein UO-44). | |||||
|
KLDC5_MOUSE
|
||||||
| NC score | 0.094018 (rank : 106) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BFQ9, Q6ZPT4 | Gene names | Klhdc5, Kiaa1340 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 5. | |||||
|
KBTBB_HUMAN
|
||||||
| NC score | 0.092014 (rank : 107) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94819 | Gene names | KBTBD11, KIAA0711, KLHDC7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch repeat and BTB domain-containing protein 11 (Kelch domain- containing protein 7B). | |||||
|
KBTBB_MOUSE
|
||||||
| NC score | 0.091247 (rank : 108) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BNW9 | Gene names | Kbtbd11, Kiaa0711 | |||
|
Domain Architecture |

|
|||||
| Description | Kelch repeat and BTB domain-containing protein 11. | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.087304 (rank : 109) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
KLD7B_HUMAN
|
||||||
| NC score | 0.086127 (rank : 110) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q96G42 | Gene names | KLHDC7B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 7B. | |||||
|
BTBD8_HUMAN
|
||||||
| NC score | 0.085762 (rank : 111) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XKL5, Q6V9S5 | Gene names | BTBD8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 8. | |||||
|
SPOP_HUMAN
|
||||||
| NC score | 0.080244 (rank : 112) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43791 | Gene names | SPOP | |||
|
Domain Architecture |

|
|||||
| Description | Speckle-type POZ protein. | |||||
|
SPOP_MOUSE
|
||||||
| NC score | 0.080244 (rank : 113) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6ZWS8 | Gene names | Spop, Pcif1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Speckle-type POZ protein (PDX-1 C-terminal-interacting factor 1) (PCIF1). | |||||
|
TMPS3_HUMAN
|
||||||
| NC score | 0.079264 (rank : 114) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.077199 (rank : 115) | θ value | 0.0431538 (rank : 39) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
RHBT3_MOUSE
|
||||||
| NC score | 0.076686 (rank : 116) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9CTN4, Q80X55, Q9CVT0 | Gene names | Rhobtb3, Kiaa0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
RHBT3_HUMAN
|
||||||
| NC score | 0.076524 (rank : 117) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O94955, Q8IW06 | Gene names | RHOBTB3, KIAA0878 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-related BTB domain-containing protein 3. | |||||
|
KLD8A_MOUSE
|
||||||
| NC score | 0.076424 (rank : 118) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q91XA8 | Gene names | Klhdc8a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8A. | |||||
|
LZTR1_HUMAN
|
||||||
| NC score | 0.076249 (rank : 119) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8N653, Q14776 | Gene names | LZTR1, TCFL2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
LZTR1_MOUSE
|
||||||
| NC score | 0.076177 (rank : 120) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9CQ33 | Gene names | Lztr1, Tcfl2 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-zipper-like transcriptional regulator 1 (LZTR-1). | |||||
|
TMPS3_MOUSE
|
||||||
| NC score | 0.075301 (rank : 121) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
ABTB1_HUMAN
|
||||||
| NC score | 0.075198 (rank : 122) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q969K4, Q6ZNU9, Q71MF1, Q96S62, Q96S63 | Gene names | ABTB1, BPOZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1 (Elongation factor 1A-binding protein). | |||||
|
TDPZ4_MOUSE
|
||||||
| NC score | 0.073560 (rank : 123) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6YCH2 | Gene names | Tdpoz4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 4. | |||||
|
KLD8A_HUMAN
|
||||||
| NC score | 0.073121 (rank : 124) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IYD2, Q9NVG5 | Gene names | KLHDC8A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8A. | |||||
|
KLD8B_MOUSE
|
||||||
| NC score | 0.072320 (rank : 125) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9D2D9 | Gene names | Klhdc8b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8B. | |||||
|
KLD8B_HUMAN
|
||||||
| NC score | 0.072287 (rank : 126) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8IXV7 | Gene names | KLHDC8B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch domain-containing protein 8B. | |||||
|
TDPZ1_MOUSE
|
||||||
| NC score | 0.070463 (rank : 127) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q717B3, Q71G58, Q8BZV7, Q8VI39 | Gene names | Tdpoz1, 2cpoz56, Mapp2, Spopl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 1 (MAPP family protein 2). | |||||
|
TDPZ2_MOUSE
|
||||||
| NC score | 0.070404 (rank : 128) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q717B2 | Gene names | Tdpoz2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 2. | |||||
|
ABTB1_MOUSE
|
||||||
| NC score | 0.069722 (rank : 129) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q99LJ2, Q8C2K7, Q91XU4 | Gene names | Abtb1, Bpoz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 1. | |||||
|
TDPZ3_MOUSE
|
||||||
| NC score | 0.065927 (rank : 130) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q717B4 | Gene names | Tdpoz3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 3. | |||||
|
TDPZ5_MOUSE
|
||||||
| NC score | 0.065922 (rank : 131) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q6YCH1 | Gene names | Tdpoz5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TD and POZ domain-containing protein 5. | |||||
|
GMCL1_MOUSE
|
||||||
| NC score | 0.065329 (rank : 132) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q920G9, Q9DBW5, Q9QUP6 | Gene names | Gmcl1, Gcl, Gcl1 | |||
|
Domain Architecture |

|
|||||
| Description | Germ cell-less protein-like 1 (mGcl-1) (DP-interacting protein) (DIP). | |||||
|
SCAR3_MOUSE
|
||||||
| NC score | 0.064426 (rank : 133) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C850, Q8C6N1 | Gene names | Scara3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3. | |||||
|
GMCL1_HUMAN
|
||||||
| NC score | 0.064190 (rank : 134) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96IK5, Q9H826, Q9H8V7, Q9H927 | Gene names | GMCL1, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Germ cell-less protein-like 1. | |||||
|
SCAR3_HUMAN
|
||||||
| NC score | 0.063154 (rank : 135) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6AZY7, Q9UM15, Q9UM16 | Gene names | SCARA3, CSR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger receptor class A member 3 (Cellular stress response gene protein). | |||||
|
BACH2_HUMAN
|
||||||
| NC score | 0.059163 (rank : 136) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9BYV9, Q59H70, Q5T793, Q9NTS5 | Gene names | BACH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
BACH2_MOUSE
|
||||||
| NC score | 0.058699 (rank : 137) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P97303 | Gene names | Bach2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH2 (BTB and CNC homolog 2). | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.058476 (rank : 138) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TMPS4_MOUSE
|
||||||
| NC score | 0.058402 (rank : 139) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
BACH1_HUMAN
|
||||||
| NC score | 0.057432 (rank : 140) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O14867, O43285 | Gene names | BACH1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1) (HA2303). | |||||
|
BACH1_MOUSE
|
||||||
| NC score | 0.057425 (rank : 141) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97302 | Gene names | Bach1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription regulator protein BACH1 (BTB and CNC homolog 1). | |||||
|
COLQ_HUMAN
|
||||||
| NC score | 0.056842 (rank : 142) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y215, Q6DK18, Q6YH18, Q6YH19, Q6YH20, Q6YH21, Q9NP18, Q9NP19, Q9NP20, Q9NP21, Q9NP22, Q9NP23, Q9NP24, Q9UP88 | Gene names | COLQ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acetylcholinesterase collagenic tail peptide precursor (AChE Q subunit) (Acetylcholinesterase-associated collagen). | |||||
|
COJA1_HUMAN
|
||||||
| NC score | 0.056791 (rank : 143) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14993, Q00559, Q05850, Q12885, Q13676, Q5JUF0, Q5T424, Q9H572, Q9NPZ2, Q9NQP2 | Gene names | COL19A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XIX) chain precursor (Collagen alpha-1(Y) chain). | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.056469 (rank : 144) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.056386 (rank : 145) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ZP2_HUMAN
|
||||||
| NC score | 0.055329 (rank : 146) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05996 | Gene names | ZP2, ZPA | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.055124 (rank : 147) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMPS5_MOUSE
|
||||||
| NC score | 0.054537 (rank : 148) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
CONA1_HUMAN
|
||||||
| NC score | 0.054524 (rank : 149) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86Y22, Q8IVR4, Q9NT93 | Gene names | COL23A1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
ABTB2_MOUSE
|
||||||
| NC score | 0.054296 (rank : 150) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7TQI7 | Gene names | Abtb2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.054180 (rank : 151) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
CONA1_MOUSE
|
||||||
| NC score | 0.053912 (rank : 152) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4G2, Q5SUQ0 | Gene names | Col23a1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXIII) chain. | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.053779 (rank : 153) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.053558 (rank : 154) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.053315 (rank : 155) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.053232 (rank : 156) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CO9A1_HUMAN
|
||||||
| NC score | 0.053168 (rank : 157) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20849, Q13699, Q13700, Q5TF52, Q6P467, Q96BM8, Q99225, Q9H151, Q9H152, Q9Y6P2, Q9Y6P3 | Gene names | COL9A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CO9A2_HUMAN
|
||||||
| NC score | 0.053041 (rank : 158) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14055 | Gene names | COL9A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.053012 (rank : 159) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.052989 (rank : 160) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.052954 (rank : 161) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CO9A1_MOUSE
|
||||||
| NC score | 0.052614 (rank : 162) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q05722, Q61269, Q61270, Q61433, Q61940 | Gene names | Col9a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IX) chain precursor. | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.052529 (rank : 163) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
GMCLL_HUMAN
|
||||||
| NC score | 0.052356 (rank : 164) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NEA9, Q8TC88, Q8TC89 | Gene names | GMCL1L, GCL | |||
|
Domain Architecture |

|
|||||
| Description | Germ cell-less protein-like 1-like. | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.052342 (rank : 165) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
TMPS7_HUMAN
|
||||||
| NC score | 0.051952 (rank : 166) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
PCOC2_MOUSE
|
||||||
| NC score | 0.051780 (rank : 167) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R4W6, Q3V1K6, Q9CX06 | Gene names | Pcolce2, Pcpe2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | 0.051685 (rank : 168) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
PCOC2_HUMAN
|
||||||
| NC score | 0.051202 (rank : 169) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKZ9, Q9BRH3 | Gene names | PCOLCE2, PCPE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Procollagen C-endopeptidase enhancer 2 precursor (Procollagen COOH- terminal proteinase enhancer 2) (Procollagen C-proteinase enhancer 2) (PCPE-2). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.050758 (rank : 170) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.050757 (rank : 171) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.050377 (rank : 172) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
ZP2_MOUSE
|
||||||
| NC score | 0.050363 (rank : 173) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20239 | Gene names | Zp2, Zp-2, Zpa | |||
|
Domain Architecture |

|
|||||
| Description | Zona pellucida sperm-binding protein 2 precursor (Zona pellucida glycoprotein ZP2) (Zona pellucida protein A). | |||||
|
CO6A1_HUMAN
|
||||||
| NC score | 0.050274 (rank : 174) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P12109, O00117, O00118, Q14040, Q14041, Q16258 | Gene names | COL6A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VI) chain precursor. | |||||
|
RPTOR_MOUSE
|
||||||
| NC score | 0.019057 (rank : 175) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K4Q0, Q8C9W9, Q8CBY4, Q8CDY8, Q9D4H3 | Gene names | Raptor | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
RPTOR_HUMAN
|
||||||
| NC score | 0.018872 (rank : 176) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N122, Q8N4V9, Q8TB32, Q9P2P3 | Gene names | RAPTOR, KIAA1303 | |||
|
Domain Architecture |

|
|||||
| Description | Regulatory-associated protein of mTOR (Raptor) (P150 target of rapamycin (TOR)-scaffold protein). | |||||
|
CNOT6_MOUSE
|
||||||
| NC score | 0.014667 (rank : 177) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K3P5, Q5NCL3, Q80V15 | Gene names | Cnot6, Ccr4, Kiaa1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
CNOT6_HUMAN
|
||||||
| NC score | 0.014185 (rank : 178) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9ULM6 | Gene names | CNOT6, CCR4, KIAA1194 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 6 (EC 3.1.-.-) (Cytoplasmic deadenylase) (Carbon catabolite repressor protein 4 homolog) (CCR4 carbon catabolite repression 4-like). | |||||
|
CTF1_MOUSE
|
||||||
| NC score | 0.012555 (rank : 179) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60753 | Gene names | Ctf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cardiotrophin-1 (CT-1). | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.010532 (rank : 180) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 67 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||