Please be patient as the page loads
|
STON1_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STON1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.987730 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON2_HUMAN
|
||||||
| θ value | 2.70778e-113 (rank : 3) | NC score | 0.906721 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 1.34386e-112 (rank : 4) | NC score | 0.917910 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
AP1M1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 5) | NC score | 0.471412 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 6) | NC score | 0.468614 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_HUMAN
|
||||||
| θ value | 7.34386e-10 (rank : 7) | NC score | 0.453156 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M2_MOUSE
|
||||||
| θ value | 9.59137e-10 (rank : 8) | NC score | 0.452435 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP2M1_HUMAN
|
||||||
| θ value | 3.64472e-09 (rank : 9) | NC score | 0.468718 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| θ value | 3.64472e-09 (rank : 10) | NC score | 0.468718 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP4M1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 11) | NC score | 0.329178 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP4M1_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 12) | NC score | 0.301846 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
ANR40_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.062858 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AI12, Q96E32 | Gene names | ANKRD40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 40. | |||||
|
ANR40_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.062176 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SUE8, Q8R595, Q9CU71 | Gene names | Ankrd40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 40. | |||||
|
BCOR_MOUSE
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.039910 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.028721 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
BCOR_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.038082 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.037738 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MTMR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.014356 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
SON_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.034251 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SON_MOUSE
|
||||||
| θ value | 2.36792 (rank : 21) | NC score | 0.034868 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TNR1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.015736 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |
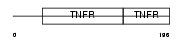
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
ANR21_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.009934 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
AP3M2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.211726 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
AP3M2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.213400 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.014582 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDX4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.008519 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
LPHN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.006230 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
POT15_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.010002 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
RAPH1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.021502 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
SETX_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.016067 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.016813 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
AP3M1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.214496 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.214366 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
DMXL1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.010166 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
FAM9A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.019045 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
P3C2G_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.008105 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.007776 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
PXK_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.015434 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7A4, Q3BCH4, Q3BCH5, Q3BCH6, Q3BDW1, Q45L83, Q59EX3, Q6PK17, Q6ZN39, Q96CA3, Q96R07, Q9NXB8 | Gene names | PXK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.020940 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.010634 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.025620 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.004259 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.011154 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.010848 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
MTMR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.011057 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.010566 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.000158 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.009265 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
STON1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON1_MOUSE
|
||||||
| NC score | 0.987730 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.917910 (rank : 3) | θ value | 1.34386e-112 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.906721 (rank : 4) | θ value | 2.70778e-113 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
AP1M1_HUMAN
|
||||||
| NC score | 0.471412 (rank : 5) | θ value | 7.09661e-13 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP2M1_HUMAN
|
||||||
| NC score | 0.468718 (rank : 6) | θ value | 3.64472e-09 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| NC score | 0.468718 (rank : 7) | θ value | 3.64472e-09 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP1M1_MOUSE
|
||||||
| NC score | 0.468614 (rank : 8) | θ value | 1.58096e-12 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_HUMAN
|
||||||
| NC score | 0.453156 (rank : 9) | θ value | 7.34386e-10 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M2_MOUSE
|
||||||
| NC score | 0.452435 (rank : 10) | θ value | 9.59137e-10 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP4M1_HUMAN
|
||||||
| NC score | 0.329178 (rank : 11) | θ value | 0.00665767 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP4M1_MOUSE
|
||||||
| NC score | 0.301846 (rank : 12) | θ value | 0.0148317 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP3M1_HUMAN
|
||||||
| NC score | 0.214496 (rank : 13) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| NC score | 0.214366 (rank : 14) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M2_MOUSE
|
||||||
| NC score | 0.213400 (rank : 15) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
AP3M2_HUMAN
|
||||||
| NC score | 0.211726 (rank : 16) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
ANR40_HUMAN
|
||||||
| NC score | 0.062858 (rank : 17) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 224 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6AI12, Q96E32 | Gene names | ANKRD40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 40. | |||||
|
ANR40_MOUSE
|
||||||
| NC score | 0.062176 (rank : 18) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q5SUE8, Q8R595, Q9CU71 | Gene names | Ankrd40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 40. | |||||
|
BCOR_MOUSE
|
||||||
| NC score | 0.039910 (rank : 19) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 521 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CGN4, Q6PDK5, Q6ZPM3, Q8BKF5, Q8CGN1, Q8CGN2, Q8CGN3 | Gene names | Bcor, Kiaa1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BCOR_HUMAN
|
||||||
| NC score | 0.038082 (rank : 20) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6W2J9, Q6P4B6, Q7Z2K7, Q8TEB4, Q96DB3, Q9H232, Q9H233, Q9HCJ7, Q9NXF2 | Gene names | BCOR, KIAA1575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BCoR protein (BCL-6 corepressor). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.037738 (rank : 21) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.034868 (rank : 22) | θ value | 2.36792 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.034251 (rank : 23) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.028721 (rank : 24) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.025620 (rank : 25) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
RAPH1_HUMAN
|
||||||
| NC score | 0.021502 (rank : 26) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q70E73, Q96Q37, Q9C0I2 | Gene names | RAPH1, ALS2CR9, KIAA1681, LPD, PREL2, RMO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-associated and pleckstrin homology domains-containing protein 1 (RAPH1) (Lamellipodin) (Proline-rich EVH1 ligand 2) (PREL-2) (Protein RMO1) (Amyotrophic lateral sclerosis 2 chromosomal region candidate 9 gene protein). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.020940 (rank : 27) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
FAM9A_HUMAN
|
||||||
| NC score | 0.019045 (rank : 28) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZU1 | Gene names | FAM9A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM9A. | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.016813 (rank : 29) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
SETX_HUMAN
|
||||||
| NC score | 0.016067 (rank : 30) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7Z333, O75120, Q3KQX4, Q68DW5, Q6AZD7, Q7Z3J6, Q8WX33, Q9H9D1, Q9NVP9 | Gene names | SETX, ALS4, KIAA0625, SCAR1 | |||
|
Domain Architecture |

|
|||||
| Description | Probable helicase senataxin (EC 3.6.1.-) (SEN1 homolog). | |||||
|
TNR1B_HUMAN
|
||||||
| NC score | 0.015736 (rank : 31) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20333, Q16042, Q6YI29, Q9UIH1 | Gene names | TNFRSF1B, TNFBR, TNFR2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor) (CD120b antigen) (Etanercept) [Contains: Tumor necrosis factor receptor superfamily member 1b, membrane form; Tumor necrosis factor-binding protein 2 (TBPII) (TBP- 2)]. | |||||
|
PXK_HUMAN
|
||||||
| NC score | 0.015434 (rank : 32) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7A4, Q3BCH4, Q3BCH5, Q3BCH6, Q3BDW1, Q45L83, Q59EX3, Q6PK17, Q6ZN39, Q96CA3, Q96R07, Q9NXB8 | Gene names | PXK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PX domain-containing protein kinase-like protein (Modulator of Na,K- ATPase) (MONaKA). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.014582 (rank : 33) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
MTMR1_MOUSE
|
||||||
| NC score | 0.014356 (rank : 34) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2C4 | Gene names | Mtmr1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.011154 (rank : 35) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
MTMR1_HUMAN
|
||||||
| NC score | 0.011057 (rank : 36) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13613, Q9UBX6, Q9UEM0, Q9UQD5 | Gene names | MTMR1 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 1 (EC 3.1.3.-). | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.010848 (rank : 37) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.010634 (rank : 38) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.010566 (rank : 39) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
DMXL1_HUMAN
|
||||||
| NC score | 0.010166 (rank : 40) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y485 | Gene names | DMXL1, XL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DmX-like 1 (X-like 1 protein). | |||||
|
POT15_HUMAN
|
||||||
| NC score | 0.010002 (rank : 41) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6S5H4, Q6NXN7, Q6S5H7 | Gene names | POTE15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prostate, ovary, testis-expressed protein on chromosome 15. | |||||
|
ANR21_HUMAN
|
||||||
| NC score | 0.009934 (rank : 42) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86YR6 | Gene names | ANKRD21, POTE | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 21 (Protein POTE) (Prostate, ovary, testis-expressed protein). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.009265 (rank : 43) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
CDX4_MOUSE
|
||||||
| NC score | 0.008519 (rank : 44) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
P3C2G_HUMAN
|
||||||
| NC score | 0.008105 (rank : 45) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75747 | Gene names | PIK3C2G | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing gamma polypeptide (EC 2.7.1.154) (Phosphoinositide 3-Kinase-C2-gamma) (PtdIns-3-kinase C2 gamma) (PI3K-C2gamma). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.007776 (rank : 46) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
LPHN1_HUMAN
|
||||||
| NC score | 0.006230 (rank : 47) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94910, Q96IE7, Q9BU07, Q9HAR3 | Gene names | LPHN1, KIAA0821, LEC2 | |||
|
Domain Architecture |

|
|||||
| Description | Latrophilin-1 precursor (Calcium-independent alpha-latrotoxin receptor 1) (Lectomedin-2). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.004259 (rank : 48) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.000158 (rank : 49) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||