Please be patient as the page loads
|
STON1_MOUSE
|
||||||
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STON1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987730 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON2_HUMAN
|
||||||
| θ value | 1.58745e-113 (rank : 3) | NC score | 0.904887 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 3.42535e-108 (rank : 4) | NC score | 0.913552 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
AP1M1_HUMAN
|
||||||
| θ value | 3.52202e-12 (rank : 5) | NC score | 0.487438 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_MOUSE
|
||||||
| θ value | 7.84624e-12 (rank : 6) | NC score | 0.484670 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP2M1_HUMAN
|
||||||
| θ value | 1.33837e-11 (rank : 7) | NC score | 0.490571 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| θ value | 1.33837e-11 (rank : 8) | NC score | 0.490571 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP1M2_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 9) | NC score | 0.470134 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M2_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 10) | NC score | 0.469441 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP4M1_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 11) | NC score | 0.322142 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 12) | NC score | 0.070440 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
AP4M1_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 13) | NC score | 0.349010 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 14) | NC score | 0.052594 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.065783 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 16) | NC score | 0.035747 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 17) | NC score | 0.047040 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
SON_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.056837 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
TCGAP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.040716 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
AP3M2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.236931 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.016944 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
NCOA6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.035570 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
AP3M1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.238178 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.238050 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.235256 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.018399 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
EPHB4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.001000 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1013 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54761, Q60627 | Gene names | Ephb4, Htk, Mdk2, Myk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-2) (Developmental kinase 2) (Tyrosine kinase MYK- 1). | |||||
|
TNR1B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.017927 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |
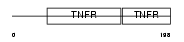
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
UBP47_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.022233 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.022101 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.025268 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
ITB5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.008087 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |
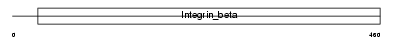
|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
SALL2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.005003 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.008526 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDX4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.008424 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
LPH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.012387 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09848 | Gene names | LCT, LPH | |||
|
Domain Architecture |

|
|||||
| Description | Lactase-phlorizin hydrolase precursor (Lactase-glycosylceramidase) [Includes: Lactase (EC 3.2.1.108); Phlorizin hydrolase (EC 3.2.1.62)]. | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.033204 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
WASL_MOUSE
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.039022 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.021298 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
PA24B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.008001 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
PRCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.040753 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.006668 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.014402 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO8A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.016574 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P25067, Q8TEJ5 | Gene names | COL8A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.017869 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
SIPA1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.007889 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.018561 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
ZN621_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | -0.001390 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZSS3, Q8TE91 | Gene names | ZNF621 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 621. | |||||
|
CO8A1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.016162 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.010261 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.052203 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
STON1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8CDJ8, Q8CDL8, Q9D5T3 | Gene names | Ston1, Salf, Sblf, Stn1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON1_HUMAN
|
||||||
| NC score | 0.987730 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y6Q2, Q96JE3, Q9BYX3 | Gene names | STON1, SALF, SBLF, STN1 | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-1 (Stoned B-like factor). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.913552 (rank : 3) | θ value | 3.42535e-108 (rank : 4) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
STON2_HUMAN
|
||||||
| NC score | 0.904887 (rank : 4) | θ value | 1.58745e-113 (rank : 3) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8WXE9, Q6NT47, Q96RI7, Q96RU6 | Gene names | STON2, STN2, STNB | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
AP2M1_HUMAN
|
||||||
| NC score | 0.490571 (rank : 5) | θ value | 1.33837e-11 (rank : 7) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96CW1, P20172, P53679 | Gene names | AP2M1, CLAPM1, KIAA0109 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (HA2 50 kDa subunit) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP2M1_MOUSE
|
||||||
| NC score | 0.490571 (rank : 6) | θ value | 1.33837e-11 (rank : 8) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P84091, P20172, P53679 | Gene names | Ap2m1, Clapm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-2 complex subunit mu-1 (Adaptin mu-1) (AP-2 mu-2 chain) (Clathrin coat assembly protein AP50) (Clathrin coat-associated protein AP50) (Plasma membrane adaptor AP-2 50 kDa protein) (Clathrin assembly protein complex 2 medium chain). | |||||
|
AP1M1_HUMAN
|
||||||
| NC score | 0.487438 (rank : 7) | θ value | 3.52202e-12 (rank : 5) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXS5 | Gene names | AP1M1, CLTNM | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M1_MOUSE
|
||||||
| NC score | 0.484670 (rank : 8) | θ value | 7.84624e-12 (rank : 6) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35585 | Gene names | Ap1m1, Cltnm | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-1 (Adaptor-related protein complex 1 mu-1 subunit) (Mu-adaptin 1) (Adaptor protein complex AP-1 mu-1 subunit) (Golgi adaptor HA1/AP1 adaptin mu-1 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 1) (AP-mu chain family member mu1A) (Clathrin coat assembly protein AP47) (Clathrin coat- associated protein AP47). | |||||
|
AP1M2_HUMAN
|
||||||
| NC score | 0.470134 (rank : 9) | θ value | 1.9326e-10 (rank : 9) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y6Q5, Q9BSI8 | Gene names | AP1M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP1M2_MOUSE
|
||||||
| NC score | 0.469441 (rank : 10) | θ value | 4.30538e-10 (rank : 10) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9WVP1, Q99LA4, Q9CWP7 | Gene names | Ap1m2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit mu-2 (Adaptor-related protein complex 1 mu-2 subunit) (Mu-adaptin 2) (Adaptor protein complex AP-1 mu-2 subunit) (Golgi adaptor HA1/AP1 adaptin mu-2 subunit) (Clathrin assembly protein assembly protein complex 1 medium chain 2) (AP-mu chain family member mu1B). | |||||
|
AP4M1_HUMAN
|
||||||
| NC score | 0.349010 (rank : 11) | θ value | 0.00665767 (rank : 13) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O00189, Q9UHK9 | Gene names | AP4M1, MUARP2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP4M1_MOUSE
|
||||||
| NC score | 0.322142 (rank : 12) | θ value | 0.00134147 (rank : 11) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC7 | Gene names | Ap4m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-4 complex subunit mu-1 (Adapter-related protein complex 4 mu-1 subunit) (Mu subunit of AP-4) (AP-4 adapter complex mu subunit) (Mu- adaptin-related protein 2) (mu-ARP2) (mu4). | |||||
|
AP3M1_HUMAN
|
||||||
| NC score | 0.238178 (rank : 13) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Y2T2 | Gene names | AP3M1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M1_MOUSE
|
||||||
| NC score | 0.238050 (rank : 14) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JKC8, Q5BKQ6 | Gene names | Ap3m1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-1 (Adapter-related protein complex 3 mu-1 subunit) (Mu-adaptin 3A) (AP-3 adapter complex mu3A subunit). | |||||
|
AP3M2_MOUSE
|
||||||
| NC score | 0.236931 (rank : 15) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8R2R9, Q3UYJ3, Q923G7 | Gene names | Ap3m2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B) (m3B). | |||||
|
AP3M2_HUMAN
|
||||||
| NC score | 0.235256 (rank : 16) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53677 | Gene names | AP3M2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit mu-2 (Adapter-related protein complex 3 mu-2 subunit) (Clathrin coat assembly protein AP47 homolog 2) (Clathrin coat-associated protein AP47 homolog 2) (Golgi adaptor AP-1 47 kDa protein homolog 2) (HA1 47 kDa subunit homolog 2) (Clathrin assembly protein assembly protein complex 1 medium chain homolog 2) (P47B). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.070440 (rank : 17) | θ value | 0.00228821 (rank : 12) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.065783 (rank : 18) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.056837 (rank : 19) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.052594 (rank : 20) | θ value | 0.00869519 (rank : 14) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.052203 (rank : 21) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.047040 (rank : 22) | θ value | 0.0252991 (rank : 17) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
PRCC_HUMAN
|
||||||
| NC score | 0.040753 (rank : 23) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92733, O00665, O00724 | Gene names | PRCC, TPRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein PRCC (Papillary renal cell carcinoma translocation-associated gene protein). | |||||
|
TCGAP_MOUSE
|
||||||
| NC score | 0.040716 (rank : 24) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80YF9, Q6P7W6 | Gene names | Snx26, Tcgap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.039022 (rank : 25) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.035747 (rank : 26) | θ value | 0.0193708 (rank : 16) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
NCOA6_HUMAN
|
||||||
| NC score | 0.035570 (rank : 27) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1202 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q14686, Q9NTZ9, Q9UH74, Q9UK86 | Gene names | NCOA6, AIB3, KIAA0181, RAP250, TRBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (PRIP) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC RAP250) (Thyroid hormone receptor-binding protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.033204 (rank : 28) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.025268 (rank : 29) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
UBP47_HUMAN
|
||||||
| NC score | 0.022233 (rank : 30) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96K76, Q658U0, Q86Y73, Q8TEP6, Q9BWI0, Q9NWN1 | Gene names | USP47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
UBP47_MOUSE
|
||||||
| NC score | 0.022101 (rank : 31) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BY87, Q5EBP2, Q6KAR9, Q80V06, Q8BHU1, Q8BI15, Q8BI16, Q8BUW4, Q91X25 | Gene names | Usp47 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 47 (EC 3.1.2.15) (Ubiquitin thioesterase 47) (Ubiquitin-specific-processing protease 47) (Deubiquitinating enzyme 47). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.021298 (rank : 32) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.018561 (rank : 33) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.018399 (rank : 34) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
TNR1B_MOUSE
|
||||||
| NC score | 0.017927 (rank : 35) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P25119, O88734, P97893 | Gene names | Tnfrsf1b, Tnfr-2, Tnfr2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1B precursor (Tumor necrosis factor receptor 2) (TNF-R2) (Tumor necrosis factor receptor type II) (p75) (p80 TNF-alpha receptor). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.017869 (rank : 36) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.016944 (rank : 37) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
CO8A2_HUMAN
|
||||||
| NC score | 0.016574 (rank : 38) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 299 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P25067, Q8TEJ5 | Gene names | COL8A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(VIII) chain precursor (Endothelial collagen). | |||||
|
CO8A1_HUMAN
|
||||||
| NC score | 0.016162 (rank : 39) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27658, Q96D07 | Gene names | COL8A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(VIII) chain precursor (Endothelial collagen). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.014402 (rank : 40) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
LPH_HUMAN
|
||||||
| NC score | 0.012387 (rank : 41) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09848 | Gene names | LCT, LPH | |||
|
Domain Architecture |

|
|||||
| Description | Lactase-phlorizin hydrolase precursor (Lactase-glycosylceramidase) [Includes: Lactase (EC 3.2.1.108); Phlorizin hydrolase (EC 3.2.1.62)]. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.010261 (rank : 42) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.008526 (rank : 43) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDX4_MOUSE
|
||||||
| NC score | 0.008424 (rank : 44) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07424 | Gene names | Cdx4, Cdx-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-4 (Caudal-type homeobox protein 4). | |||||
|
ITB5_MOUSE
|
||||||
| NC score | 0.008087 (rank : 45) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70309, O70308, O88347 | Gene names | Itgb5 | |||
|
Domain Architecture |

|
|||||
| Description | Integrin beta-5 precursor. | |||||
|
PA24B_MOUSE
|
||||||
| NC score | 0.008001 (rank : 46) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4QQM1, Q80VV8, Q91W88 | Gene names | Pla2g4b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytosolic phospholipase A2 beta (EC 3.1.1.4) (cPLA2-beta) (Phospholipase A2 group IVB). | |||||
|
SIPA1_MOUSE
|
||||||
| NC score | 0.007889 (rank : 47) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P46062, P70204 | Gene names | Sipa1, Spa-1, Spa1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-induced proliferation-associated protein 1 (Sipa-1) (GTPase- activating protein Spa-1). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.006668 (rank : 48) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
SALL2_HUMAN
|
||||||
| NC score | 0.005003 (rank : 49) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y467, Q9Y4G1 | Gene names | SALL2, KIAA0360, SAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sal-like protein 2 (Zinc finger protein SALL2) (HSal2). | |||||
|
EPHB4_MOUSE
|
||||||
| NC score | 0.001000 (rank : 50) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 1013 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54761, Q60627 | Gene names | Ephb4, Htk, Mdk2, Myk1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-B receptor 4 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor MDK-2) (Developmental kinase 2) (Tyrosine kinase MYK- 1). | |||||
|
ZN621_HUMAN
|
||||||
| NC score | -0.001390 (rank : 51) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 50 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZSS3, Q8TE91 | Gene names | ZNF621 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 621. | |||||