Please be patient as the page loads
|
CHRC1_HUMAN
|
||||||
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CHRC1_HUMAN
|
||||||
| θ value | 2.41096e-53 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
CHRC1_MOUSE
|
||||||
| θ value | 9.18288e-45 (rank : 2) | NC score | 0.982231 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKP8, Q91VG2 | Gene names | Chrac1 | |||
|
Domain Architecture |
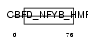
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (DNA polymerase epsilon subunit p15) (YC-like protein 1) (YCL1) (NF-YC-like protein). | |||||
|
NFYC_HUMAN
|
||||||
| θ value | 1.38499e-08 (rank : 3) | NC score | 0.609406 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
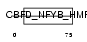
Domain Architecture |
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
NFYC_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 4) | NC score | 0.652683 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
DPOE4_MOUSE
|
||||||
| θ value | 4.0297e-08 (rank : 5) | NC score | 0.703810 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |
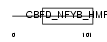
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DPOE4_HUMAN
|
||||||
| θ value | 6.87365e-08 (rank : 6) | NC score | 0.696595 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR33, Q53TR2 | Gene names | POLE4 | |||
|
Domain Architecture |
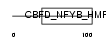
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DRAP1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.417894 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14919, Q13448 | Gene names | DRAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
DRAP1_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 8) | NC score | 0.405346 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
USP9X_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.069656 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9X_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.068156 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
SAMN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.024979 (rank : 14) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSI8, Q8NFF7, Q9C041 | Gene names | SAMSN1, HACS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain-containing protein SAMSN-1 (SAM domain, SH3 domain and nuclear localization signals protein 1) (SH3-SAM adaptor protein) (Hematopoietic adaptor containing SH3 and SAM domains 1). | |||||
|
USP9Y_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.062447 (rank : 13) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
DPOE3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 13) | NC score | 0.106537 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
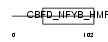
Domain Architecture |
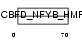
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
DPOE3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.112083 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
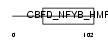
Domain Architecture |
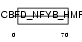
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
CHRC1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.41096e-53 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
CHRC1_MOUSE
|
||||||
| NC score | 0.982231 (rank : 2) | θ value | 9.18288e-45 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JKP8, Q91VG2 | Gene names | Chrac1 | |||
|
Domain Architecture |
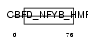
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (DNA polymerase epsilon subunit p15) (YC-like protein 1) (YCL1) (NF-YC-like protein). | |||||
|
DPOE4_MOUSE
|
||||||
| NC score | 0.703810 (rank : 3) | θ value | 4.0297e-08 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DPOE4_HUMAN
|
||||||
| NC score | 0.696595 (rank : 4) | θ value | 6.87365e-08 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NR33, Q53TR2 | Gene names | POLE4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
NFYC_MOUSE
|
||||||
| NC score | 0.652683 (rank : 5) | θ value | 1.38499e-08 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
NFYC_HUMAN
|
||||||
| NC score | 0.609406 (rank : 6) | θ value | 1.38499e-08 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
DRAP1_HUMAN
|
||||||
| NC score | 0.417894 (rank : 7) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14919, Q13448 | Gene names | DRAP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
DRAP1_MOUSE
|
||||||
| NC score | 0.405346 (rank : 8) | θ value | 0.00869519 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D6N5 | Gene names | Drap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dr1-associated corepressor (Dr1-associated protein 1) (Negative co- factor 2 alpha) (NC2 alpha). | |||||
|
DPOE3_MOUSE
|
||||||
| NC score | 0.112083 (rank : 9) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
DPOE3_HUMAN
|
||||||
| NC score | 0.106537 (rank : 10) | θ value | θ > 10 (rank : 13) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
USP9X_MOUSE
|
||||||
| NC score | 0.069656 (rank : 11) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70398, Q62497 | Gene names | Usp9x, Fafl, Fam | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome) (Ubiquitin carboxyl-terminal hydrolase FAM) (Fat facets homolog). | |||||
|
USP9X_HUMAN
|
||||||
| NC score | 0.068156 (rank : 12) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q93008, O75550, Q8WWT3, Q8WX12 | Gene names | USP9X, DFFRX, USP9 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-X (EC 3.1.2.15) (Ubiquitin thioesterase FAF-X) (Ubiquitin-specific-processing protease FAF-X) (Deubiquitinating enzyme FAF-X) (Fat facets protein-related, X- linked) (Ubiquitin-specific protease 9, X chromosome). | |||||
|
USP9Y_HUMAN
|
||||||
| NC score | 0.062447 (rank : 13) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00507, O14601 | Gene names | USP9Y, DFFRY, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.1.2.15) (Ubiquitin thioesterase FAF-Y) (Ubiquitin-specific-processing protease FAF-Y) (Deubiquitinating enzyme FAF-Y) (Fat facets protein-related, Y- linked) (Ubiquitin-specific protease 9, Y chromosome). | |||||
|
SAMN1_HUMAN
|
||||||
| NC score | 0.024979 (rank : 14) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NSI8, Q8NFF7, Q9C041 | Gene names | SAMSN1, HACS1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain-containing protein SAMSN-1 (SAM domain, SH3 domain and nuclear localization signals protein 1) (SH3-SAM adaptor protein) (Hematopoietic adaptor containing SH3 and SAM domains 1). | |||||