Please be patient as the page loads
|
DPOE3_HUMAN
|
||||||
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |
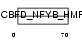
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DPOE3_MOUSE
|
||||||
| θ value | 8.30557e-46 (rank : 1) | NC score | 0.977565 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |
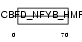
|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
DPOE3_HUMAN
|
||||||
| θ value | 1.08474e-45 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
NFYB_HUMAN
|
||||||
| θ value | 3.18553e-13 (rank : 3) | NC score | 0.721893 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25208, Q96IY8 | Gene names | NFYB, HAP3 | |||
|
Domain Architecture |
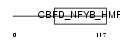
|
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
NFYB_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 4) | NC score | 0.721882 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63139, P22569, Q3UK54 | Gene names | Nfyb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
TBAP_HUMAN
|
||||||
| θ value | 6.21693e-09 (rank : 5) | NC score | 0.675340 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01658 | Gene names | DR1 | |||
|
Domain Architecture |
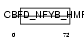
|
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Dr1 protein) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
TBAP_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 6) | NC score | 0.673168 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WV0, Q3UT14 | Gene names | Dr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
CCD16_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.197477 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
CCD16_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.196485 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
CHRC1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.220639 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKP8, Q91VG2 | Gene names | Chrac1 | |||
|
Domain Architecture |
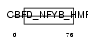
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (DNA polymerase epsilon subunit p15) (YC-like protein 1) (YCL1) (NF-YC-like protein). | |||||
|
MATR3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 10) | NC score | 0.137557 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
SEC63_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.099679 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
DPOE4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 12) | NC score | 0.224953 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR33, Q53TR2 | Gene names | POLE4 | |||
|
Domain Architecture |
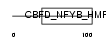
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DPOE4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.225717 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |
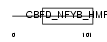
|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.080758 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
MATR3_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.139858 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
CNTRB_MOUSE
|
||||||
| θ value | 0.279714 (rank : 16) | NC score | 0.058087 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
SEC63_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.102281 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
OPTN_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.048991 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.075170 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.061826 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.064547 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
P52K_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.047649 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
PRG3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.034542 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.061103 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.034769 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.036113 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
NFYC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.139917 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |
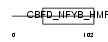
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
NFYC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.153193 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |
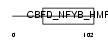
|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.039057 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.039401 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MYH6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.031232 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.034191 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
F10A1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.021020 (rank : 59) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
KIF3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.027187 (rank : 55) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
KIF3A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.028295 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.023796 (rank : 56) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.037905 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ZBTB6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.002185 (rank : 63) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |
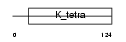
|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.033834 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.032427 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
NEK1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.002564 (rank : 62) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.006290 (rank : 61) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.030875 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.041805 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.042378 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.023699 (rank : 57) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.035367 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.034971 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
TXLNB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.032672 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.030149 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.050862 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.027981 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.020978 (rank : 60) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
S6OS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.022196 (rank : 58) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1H7 | Gene names | SIX6OS1, C14orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SIX6OS1 (Six6 opposite strand transcript 1). | |||||
|
CHRC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.106537 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |
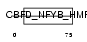
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.069219 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.057250 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.093326 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.086413 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.101128 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.087995 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RBM25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.052503 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.061859 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
DPOE3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.08474e-45 (rank : 2) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9NRF9, Q5W0U1, Q8N758, Q8NCE5, Q9NR32 | Gene names | POLE3, CHRAC17 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (Chromatin accessibility complex 17) (HuCHRAC17) (CHRAC-17) (Arsenic- transactivated protein) (AsTP). | |||||
|
DPOE3_MOUSE
|
||||||
| NC score | 0.977565 (rank : 2) | θ value | 8.30557e-46 (rank : 1) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JKP7, Q8R198 | Gene names | Pole3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 3 (EC 2.7.7.7) (DNA polymerase II subunit 3) (DNA polymerase epsilon subunit p17) (YB-like protein 1) (YBL1) (NF-YB-like protein). | |||||
|
NFYB_HUMAN
|
||||||
| NC score | 0.721893 (rank : 3) | θ value | 3.18553e-13 (rank : 3) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P25208, Q96IY8 | Gene names | NFYB, HAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
NFYB_MOUSE
|
||||||
| NC score | 0.721882 (rank : 4) | θ value | 3.18553e-13 (rank : 4) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P63139, P22569, Q3UK54 | Gene names | Nfyb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear transcription factor Y subunit beta (Nuclear transcription factor Y subunit B) (NF-YB) (CAAT-box DNA-binding protein subunit B). | |||||
|
TBAP_HUMAN
|
||||||
| NC score | 0.675340 (rank : 5) | θ value | 6.21693e-09 (rank : 5) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01658 | Gene names | DR1 | |||
|
Domain Architecture |

|
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Dr1 protein) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
TBAP_MOUSE
|
||||||
| NC score | 0.673168 (rank : 6) | θ value | 6.21693e-09 (rank : 6) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91WV0, Q3UT14 | Gene names | Dr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA-binding protein-associated phosphoprotein (Down-regulator of transcription 1) (Negative cofactor 2 beta) (NC2 beta). | |||||
|
DPOE4_MOUSE
|
||||||
| NC score | 0.225717 (rank : 7) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CQ36, Q3TJ13 | Gene names | Pole4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
DPOE4_HUMAN
|
||||||
| NC score | 0.224953 (rank : 8) | θ value | 0.0961366 (rank : 12) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NR33, Q53TR2 | Gene names | POLE4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase epsilon subunit 4 (EC 2.7.7.7) (DNA polymerase II subunit 4) (DNA polymerase epsilon subunit p12). | |||||
|
CHRC1_MOUSE
|
||||||
| NC score | 0.220639 (rank : 9) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JKP8, Q91VG2 | Gene names | Chrac1 | |||
|
Domain Architecture |
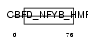
|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (DNA polymerase epsilon subunit p15) (YC-like protein 1) (YCL1) (NF-YC-like protein). | |||||
|
CCD16_HUMAN
|
||||||
| NC score | 0.197477 (rank : 10) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q96NB3, Q96F60, Q96GZ5, Q9BU38 | Gene names | CCDC16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16. | |||||
|
CCD16_MOUSE
|
||||||
| NC score | 0.196485 (rank : 11) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8R1N0, Q3TR52, Q9CWV9, Q9CYI6 | Gene names | Ccdc16, Omcg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 16 (Ovus mutant candidate gene 1 protein). | |||||
|
NFYC_MOUSE
|
||||||
| NC score | 0.153193 (rank : 12) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P70353, O35088, Q6GU21 | Gene names | Nfyc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C). | |||||
|
NFYC_HUMAN
|
||||||
| NC score | 0.139917 (rank : 13) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13952, Q5T6K8, Q5T6L1, Q5TZR6, Q92869, Q9HBX1, Q9NXB5, Q9UM67, Q9UML0, Q9UMT7 | Gene names | NFYC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear transcription factor Y subunit gamma (Nuclear transcription factor Y subunit C) (NF-YC) (CAAT-box DNA-binding protein subunit C) (Transactivator HSM-1/2). | |||||
|
MATR3_HUMAN
|
||||||
| NC score | 0.139858 (rank : 14) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43243, Q9UHW0, Q9UQ27 | Gene names | MATR3, KIAA0723 | |||
|
Domain Architecture |

|
|||||
| Description | Matrin-3. | |||||
|
MATR3_MOUSE
|
||||||
| NC score | 0.137557 (rank : 15) | θ value | 0.0431538 (rank : 10) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8K310 | Gene names | Matr3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Matrin-3. | |||||
|
CHRC1_HUMAN
|
||||||
| NC score | 0.106537 (rank : 16) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRG0 | Gene names | CHRAC1, CHRAC15 | |||
|
Domain Architecture |

|
|||||
| Description | Chromatin accessibility complex protein 1 (CHRAC-1) (CHRAC-15) (HuCHRAC15) (DNA polymerase epsilon subunit p15). | |||||
|
SEC63_MOUSE
|
||||||
| NC score | 0.102281 (rank : 17) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 601 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8VHE0, Q8VEB9 | Gene names | Sec63, Sec63l | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.101128 (rank : 18) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
SEC63_HUMAN
|
||||||
| NC score | 0.099679 (rank : 19) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UGP8, O95380, Q86VS9, Q8IWL0, Q9NTE0 | Gene names | SEC63, SEC63L | |||
|
Domain Architecture |

|
|||||
| Description | Translocation protein SEC63 homolog. | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.093326 (rank : 20) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.087995 (rank : 21) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.086413 (rank : 22) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.080758 (rank : 23) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.075170 (rank : 24) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.069219 (rank : 25) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.064547 (rank : 26) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.061859 (rank : 27) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.061826 (rank : 28) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.061103 (rank : 29) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
CNTRB_MOUSE
|
||||||
| NC score | 0.058087 (rank : 30) | θ value | 0.279714 (rank : 16) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CB62, Q5NCF3 | Gene names | Cntrob, Lip8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrobin (LYST-interacting protein 8). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.057250 (rank : 31) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.052503 (rank : 32) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.050862 (rank : 33) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.048991 (rank : 34) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
P52K_HUMAN
|
||||||
| NC score | 0.047649 (rank : 35) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43422, Q8WTW1, Q9Y3Z4 | Gene names | PRKRIR, DAP4, P52RIPK, THAP0 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa repressor of the inhibitor of the protein kinase (p58IPK- interacting protein) (58 kDa interferon-induced protein kinase- interacting protein) (P52rIPK) (Death-associated protein 4) (THAP domain-containing protein 0). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.042378 (rank : 36) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.041805 (rank : 37) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.039401 (rank : 38) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.039057 (rank : 39) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.037905 (rank : 40) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.036113 (rank : 41) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.035367 (rank : 42) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.034971 (rank : 43) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.034769 (rank : 44) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
PRG3_MOUSE
|
||||||
| NC score | 0.034542 (rank : 45) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JL95, Q3SXA9 | Gene names | Prg3, Mbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan 3 precursor (Eosinophil major basic protein 2). | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.034191 (rank : 46) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.033834 (rank : 47) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
TXLNB_HUMAN
|
||||||
| NC score | 0.032672 (rank : 48) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 912 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N3L3, Q76L25, Q86T52, Q8N3S2 | Gene names | TXLNB, C6orf198, MDP77 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Beta-taxilin (Muscle-derived protein 77) (hMDP77). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.032427 (rank : 49) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH6_MOUSE
|
||||||
| NC score | 0.031232 (rank : 50) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1501 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02566, Q64258, Q64738 | Gene names | Myh6, Myhca | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.030875 (rank : 51) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.030149 (rank : 52) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
KIF3A_MOUSE
|
||||||
| NC score | 0.028295 (rank : 53) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 715 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P28741 | Gene names | Kif3a, Kif3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.027981 (rank : 54) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
KIF3A_HUMAN
|
||||||
| NC score | 0.027187 (rank : 55) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y496, Q86XE9, Q9Y6V4 | Gene names | KIF3A, KIF3 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF3A (Microtubule plus end-directed kinesin motor 3A). | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.023796 (rank : 56) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.023699 (rank : 57) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
S6OS1_HUMAN
|
||||||
| NC score | 0.022196 (rank : 58) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8N1H7 | Gene names | SIX6OS1, C14orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein SIX6OS1 (Six6 opposite strand transcript 1). | |||||
|
F10A1_HUMAN
|
||||||
| NC score | 0.021020 (rank : 59) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P50502, O14999 | Gene names | ST13, FAM10A1, HIP, SNC6 | |||
|
Domain Architecture |

|
|||||
| Description | Hsc70-interacting protein (Hip) (Suppression of tumorigenicity protein 13) (Putative tumor suppressor ST13) (Protein FAM10A1) (Progesterone receptor-associated p48 protein) (NY-REN-33 antigen). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.020978 (rank : 60) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.006290 (rank : 61) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
NEK1_MOUSE
|
||||||
| NC score | 0.002564 (rank : 62) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P51954 | Gene names | Nek1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek1 (EC 2.7.11.1) (NimA-related protein kinase 1). | |||||
|
ZBTB6_HUMAN
|
||||||
| NC score | 0.002185 (rank : 63) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 54 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15916 | Gene names | ZBTB6, ZID, ZNF482 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 6 (Zinc finger protein 482) (Zinc finger protein with interaction domain). | |||||