Please be patient as the page loads
|
GASP1_HUMAN
|
||||||
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GASP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.951839 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 3.97187e-173 (rank : 3) | NC score | 0.934740 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 8.59026e-160 (rank : 4) | NC score | 0.909777 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
ARMX5_HUMAN
|
||||||
| θ value | 6.35935e-46 (rank : 5) | NC score | 0.850730 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1M9, Q68DB4, Q9BVZ3, Q9H969 | Gene names | ARMCX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX5_MOUSE
|
||||||
| θ value | 3.85808e-43 (rank : 6) | NC score | 0.836106 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 7) | NC score | 0.771674 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 6.38894e-30 (rank : 8) | NC score | 0.765609 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 8.3442e-30 (rank : 9) | NC score | 0.751618 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 10) | NC score | 0.752428 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 2.35151e-24 (rank : 11) | NC score | 0.705379 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 3.07116e-24 (rank : 12) | NC score | 0.661379 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ARMX4_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 13) | NC score | 0.435556 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5H9R4, Q5H9K8, Q8N8D6 | Gene names | ARMCX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 4. | |||||
|
ARMX6_HUMAN
|
||||||
| θ value | 9.29e-05 (rank : 14) | NC score | 0.509849 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7L4S7, Q9NWJ3 | Gene names | ARMCX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 15) | NC score | 0.058091 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 16) | NC score | 0.071586 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ARMX6_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 17) | NC score | 0.464213 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3A6, Q3TUH4 | Gene names | Armcx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
MYST3_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 18) | NC score | 0.029812 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 19) | NC score | 0.075876 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 20) | NC score | 0.054997 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 21) | NC score | 0.047504 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 22) | NC score | 0.086079 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
FUSIP_HUMAN
|
||||||
| θ value | 0.125558 (rank : 23) | NC score | 0.079963 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |
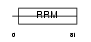
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| θ value | 0.125558 (rank : 24) | NC score | 0.079963 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |
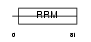
|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
HNRPU_HUMAN
|
||||||
| θ value | 0.125558 (rank : 25) | NC score | 0.036862 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 0.21417 (rank : 26) | NC score | 0.029104 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.279714 (rank : 27) | NC score | 0.012892 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 28) | NC score | 0.112881 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SPT6H_MOUSE
|
||||||
| θ value | 0.365318 (rank : 29) | NC score | 0.032482 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
DGLB_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.028111 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NCG7, Q6PIX3, Q8N2N2, Q8N9S1, Q8TED3, Q8WXE6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta) (KCCR13L). | |||||
|
AN32C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 31) | NC score | 0.021951 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43423 | Gene names | ANP32C, PP32R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C (Phosphoprotein 32-related protein 1) (Tumorigenic protein pp32r1). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.056141 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
LYRIC_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.031427 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.032406 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.029583 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CENPC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.047131 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
MECP2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.047367 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |
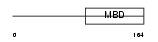
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
TCF8_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.002168 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.016597 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
CT116_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.022817 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96HY6, Q9BW47 | Gene names | C20orf116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 precursor. | |||||
|
GP152_MOUSE
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.022855 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
MECP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.043306 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |
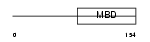
|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.064018 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
PCOC1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.015819 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
WDR43_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.017000 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
GP152_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.017364 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
IRX3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.013675 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.006779 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
RBMX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.028240 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |
|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
AHTF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.020217 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
AIOL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.000678 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKT9, Q8WWQ9, Q8WWR0, Q8WWR1, Q8WWR2, Q8WWR3 | Gene names | IKZF3, ZNFN1A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
ERBB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | -0.000177 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
LMO7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.016131 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
UBP35_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.011851 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
ANLN_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.019688 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.018004 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.047224 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RILP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.018486 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
RPA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.012247 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.010449 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
ULE1B_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.017066 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.008940 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
CENPB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.020919 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CMGA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.018283 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
RIMB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.008445 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
RIOK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.013857 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.041258 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TRI26_HUMAN
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.003537 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12899 | Gene names | TRIM26, RNF95, ZNF173 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 26 (Zinc finger protein 173) (Acid finger protein) (AFP) (RING finger protein 95). | |||||
|
CENPB_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.017918 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
HMGA1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.051199 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17096, P10910, Q9UKB0 | Gene names | HMGA1, HMGIY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.020518 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.022097 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
ULE1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.015511 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.028518 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
IRX3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.009828 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.009236 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
NOL11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.009881 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8H0, Q7L5S1, Q9UG18 | Gene names | NOL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 11. | |||||
|
NOL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.014751 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.025468 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
ROR1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | -0.002027 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z139 | Gene names | Ror1, Ntrkr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1) (mROR1). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.047880 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
ZRAN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.009352 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.050070 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF081_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.130455 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
CF081_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.105180 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050777 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.054300 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFRS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.051090 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP1_MOUSE
|
||||||
| NC score | 0.951839 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.934740 (rank : 3) | θ value | 3.97187e-173 (rank : 3) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.909777 (rank : 4) | θ value | 8.59026e-160 (rank : 4) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
ARMX5_HUMAN
|
||||||
| NC score | 0.850730 (rank : 5) | θ value | 6.35935e-46 (rank : 5) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1M9, Q68DB4, Q9BVZ3, Q9H969 | Gene names | ARMCX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX5_MOUSE
|
||||||
| NC score | 0.836106 (rank : 6) | θ value | 3.85808e-43 (rank : 6) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.771674 (rank : 7) | θ value | 1.98606e-31 (rank : 7) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.765609 (rank : 8) | θ value | 6.38894e-30 (rank : 8) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.752428 (rank : 9) | θ value | 2.42779e-29 (rank : 10) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.751618 (rank : 10) | θ value | 8.3442e-30 (rank : 9) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.705379 (rank : 11) | θ value | 2.35151e-24 (rank : 11) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.661379 (rank : 12) | θ value | 3.07116e-24 (rank : 12) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ARMX6_HUMAN
|
||||||
| NC score | 0.509849 (rank : 13) | θ value | 9.29e-05 (rank : 14) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7L4S7, Q9NWJ3 | Gene names | ARMCX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX6_MOUSE
|
||||||
| NC score | 0.464213 (rank : 14) | θ value | 0.0113563 (rank : 17) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3A6, Q3TUH4 | Gene names | Armcx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX4_HUMAN
|
||||||
| NC score | 0.435556 (rank : 15) | θ value | 4.1701e-05 (rank : 13) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5H9R4, Q5H9K8, Q8N8D6 | Gene names | ARMCX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 4. | |||||
|
CF081_HUMAN
|
||||||
| NC score | 0.130455 (rank : 16) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.112881 (rank : 17) | θ value | 0.279714 (rank : 28) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
CF081_MOUSE
|
||||||
| NC score | 0.105180 (rank : 18) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.086079 (rank : 19) | θ value | 0.0961366 (rank : 22) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
FUSIP_HUMAN
|
||||||
| NC score | 0.079963 (rank : 20) | θ value | 0.125558 (rank : 23) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O75494, O60572, Q96G09, Q96P17 | Gene names | FUSIP1, TASR | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (40 kDa SR-repressor protein) (SRrp40) (Splicing factor SRp38). | |||||
|
FUSIP_MOUSE
|
||||||
| NC score | 0.079963 (rank : 21) | θ value | 0.125558 (rank : 24) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9R0U0, O70307, O88468, Q3TGW7, Q8CFZ1, Q9R0T9 | Gene names | Fusip1, Nssr | |||
|
Domain Architecture |

|
|||||
| Description | FUS-interacting serine-arginine-rich protein 1 (TLS-associated protein with Ser-Arg repeats) (TLS-associated protein with SR repeats) (TASR) (TLS-associated serine-arginine protein) (TLS-associated SR protein) (Neural-specific SR protein) (Neural-salient serine/arginine-rich protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.075876 (rank : 22) | θ value | 0.0431538 (rank : 19) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.071586 (rank : 23) | θ value | 0.00228821 (rank : 16) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.064018 (rank : 24) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.058091 (rank : 25) | θ value | 0.00175202 (rank : 15) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.056141 (rank : 26) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.054997 (rank : 27) | θ value | 0.0736092 (rank : 20) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.054300 (rank : 28) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
HMGA1_HUMAN
|
||||||
| NC score | 0.051199 (rank : 29) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17096, P10910, Q9UKB0 | Gene names | HMGA1, HMGIY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | High mobility group protein HMG-I/HMG-Y (HMG-I(Y)) (High mobility group AT-hook protein 1) (High mobility group protein A1) (High mobility group protein-R). | |||||
|
SFRS6_HUMAN
|
||||||
| NC score | 0.051090 (rank : 30) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13247, Q13244, Q13245, Q96J06, Q9UJB8, Q9Y3N7 | Gene names | SFRS6, SRP55 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 6 (Pre-mRNA-splicing factor SRP55). | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.050777 (rank : 31) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.050070 (rank : 32) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.047880 (rank : 33) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.047504 (rank : 34) | θ value | 0.0961366 (rank : 21) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MECP2_MOUSE
|
||||||
| NC score | 0.047367 (rank : 35) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2D6 | Gene names | Mecp2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.047224 (rank : 36) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
CENPC_HUMAN
|
||||||
| NC score | 0.047131 (rank : 37) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q03188, Q9P0M5 | Gene names | CENPC1, CENPC, ICEN7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein C 1 (CENP-C) (Centromere autoantigen C) (Interphase centromere complex protein 7). | |||||
|
MECP2_HUMAN
|
||||||
| NC score | 0.043306 (rank : 38) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P51608, O15233 | Gene names | MECP2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyl-CpG-binding protein 2 (MeCP-2 protein) (MeCP2). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.041258 (rank : 39) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
HNRPU_HUMAN
|
||||||
| NC score | 0.036862 (rank : 40) | θ value | 0.125558 (rank : 25) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q00839, O75507, Q96HY9 | Gene names | HNRPU, SAFA, U21.1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Scaffold attachment factor A) (SAF-A) (p120) (pp120). | |||||
|
SPT6H_MOUSE
|
||||||
| NC score | 0.032482 (rank : 41) | θ value | 0.365318 (rank : 29) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62383, Q5SYM0, Q6GQS3, Q6ZQI0, Q8BQY6 | Gene names | Supt6h, Kiaa0162 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT6. | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.032406 (rank : 42) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
LYRIC_HUMAN
|
||||||
| NC score | 0.031427 (rank : 43) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UE4, Q6PK07, Q8TCX3 | Gene names | MTDH, AEG1, LYRIC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LYRIC (Lysine-rich CEACAM1 co-isolated protein) (3D3/lyric) (Metastasis adhesion protein) (Metadherin) (Astrocyte elevated gene-1 protein) (AEG-1). | |||||
|
MYST3_MOUSE
|
||||||
| NC score | 0.029812 (rank : 44) | θ value | 0.0431538 (rank : 18) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 688 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BZ21, Q8BW52, Q8BYH1, Q8C1F3 | Gene names | Myst3, Moz | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Monocytic leukemia zinc finger protein) (Monocytic leukemia zinc finger homolog). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.029583 (rank : 45) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.029104 (rank : 46) | θ value | 0.21417 (rank : 26) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.028518 (rank : 47) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
RBMX2_HUMAN
|
||||||
| NC score | 0.028240 (rank : 48) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y388, Q5JY82, Q9Y3I8 | Gene names | RBMX2 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding motif protein, X-linked 2. | |||||
|
DGLB_HUMAN
|
||||||
| NC score | 0.028111 (rank : 49) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NCG7, Q6PIX3, Q8N2N2, Q8N9S1, Q8TED3, Q8WXE6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta) (KCCR13L). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.025468 (rank : 50) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
GP152_MOUSE
|
||||||
| NC score | 0.022855 (rank : 51) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BXS7 | Gene names | Gpr152 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152. | |||||
|
CT116_HUMAN
|
||||||
| NC score | 0.022817 (rank : 52) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96HY6, Q9BW47 | Gene names | C20orf116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf116 precursor. | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.022097 (rank : 53) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
AN32C_HUMAN
|
||||||
| NC score | 0.021951 (rank : 54) | θ value | 0.62314 (rank : 31) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43423 | Gene names | ANP32C, PP32R1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member C (Phosphoprotein 32-related protein 1) (Tumorigenic protein pp32r1). | |||||
|
CENPB_HUMAN
|
||||||
| NC score | 0.020919 (rank : 55) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P07199 | Gene names | CENPB | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.020518 (rank : 56) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
AHTF1_HUMAN
|
||||||
| NC score | 0.020217 (rank : 57) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8WYP5, Q7Z4E3, Q8IZA4, Q96EH9, Q9Y4Q6 | Gene names | AHCTF1, ELYS, TMBS62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-hook-containing transcription factor 1 (Embryonic large molecule derived from yolk sac). | |||||
|
ANLN_MOUSE
|
||||||
| NC score | 0.019688 (rank : 58) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K298, Q8BL79, Q8BLB3, Q8K2N0, Q9CUY0 | Gene names | Anln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
RILP_HUMAN
|
||||||
| NC score | 0.018486 (rank : 59) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96NA2, Q71RE6, Q9BSL3, Q9BYS3 | Gene names | RILP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab-interacting lysosomal protein. | |||||
|
CMGA_MOUSE
|
||||||
| NC score | 0.018283 (rank : 60) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P26339 | Gene names | Chga | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) [Contains: Pancreastatin; Beta-granin; WE-14]. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.018004 (rank : 61) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.017918 (rank : 62) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
GP152_HUMAN
|
||||||
| NC score | 0.017364 (rank : 63) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TDT2, Q86SM0 | Gene names | GPR152, PGR5 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 152 (G-protein coupled receptor PGR5). | |||||
|
ULE1B_MOUSE
|
||||||
| NC score | 0.017066 (rank : 64) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1F9, Q8BVX9 | Gene names | Uble1b, Sae2, Uba2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
WDR43_HUMAN
|
||||||
| NC score | 0.017000 (rank : 65) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15061, Q15395, Q92577 | Gene names | WDR43, KIAA0007 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 43. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.016597 (rank : 66) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
LMO7_HUMAN
|
||||||
| NC score | 0.016131 (rank : 67) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WWI1, O15462, O95346, Q9UKC1, Q9UQM5, Q9Y6A7 | Gene names | LMO7, FBX20, FBXO20, KIAA0858 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain only protein 7 (LOMP) (F-box only protein 20). | |||||
|
PCOC1_MOUSE
|
||||||
| NC score | 0.015819 (rank : 68) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61398, O35113 | Gene names | Pcolce, Pcpe1 | |||
|
Domain Architecture |

|
|||||
| Description | Procollagen C-endopeptidase enhancer 1 precursor (Procollagen COOH- terminal proteinase enhancer 1) (Procollagen C-proteinase enhancer 1) (PCPE-1) (Type I procollagen COOH-terminal proteinase enhancer) (Type 1 procollagen C-proteinase enhancer protein) (P14). | |||||
|
ULE1B_HUMAN
|
||||||
| NC score | 0.015511 (rank : 69) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UBT2, O95605, Q9NTJ1, Q9UED2 | Gene names | UBLE1B, SAE2, UBA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-like 1-activating enzyme E1B (SUMO-1-activating enzyme subunit 2) (Anthracycline-associated resistance ARX). | |||||
|
NOL1_HUMAN
|
||||||
| NC score | 0.014751 (rank : 70) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46087 | Gene names | NOL1 | |||
|
Domain Architecture |

|
|||||
| Description | Proliferating-cell nucleolar antigen p120 (Proliferation-associated nucleolar protein p120). | |||||
|
RIOK2_MOUSE
|
||||||
| NC score | 0.013857 (rank : 71) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQS5, Q91XF3 | Gene names | Riok2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO2 (EC 2.7.11.1) (RIO kinase 2). | |||||
|
IRX3_HUMAN
|
||||||
| NC score | 0.013675 (rank : 72) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.012892 (rank : 73) | θ value | 0.279714 (rank : 27) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RPA1_MOUSE
|
||||||
| NC score | 0.012247 (rank : 74) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35134, Q7TSA9 | Gene names | Polr1a, Rpa1, Rpo1-4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase I largest subunit (EC 2.7.7.6) (RNA polymerase I 194 kDa subunit) (RPA194). | |||||
|
UBP35_HUMAN
|
||||||
| NC score | 0.011851 (rank : 75) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2H5 | Gene names | USP35, KIAA1372, USP34 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 35 (EC 3.1.2.15) (Ubiquitin thioesterase 35) (Ubiquitin-specific-processing protease 35) (Deubiquitinating enzyme 35). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.010449 (rank : 76) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
NOL11_HUMAN
|
||||||
| NC score | 0.009881 (rank : 77) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8H0, Q7L5S1, Q9UG18 | Gene names | NOL11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein 11. | |||||
|
IRX3_MOUSE
|
||||||
| NC score | 0.009828 (rank : 78) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
ZRAN1_HUMAN
|
||||||
| NC score | 0.009352 (rank : 79) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UGI0 | Gene names | ZRANB1, TRABID | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger Ran-binding domain-containing protein 1 (Protein TRABID). | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.009236 (rank : 80) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.008940 (rank : 81) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
RIMB2_HUMAN
|
||||||
| NC score | 0.008445 (rank : 82) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15034, Q96ID2 | Gene names | RIMBP2, KIAA0318, RBP2 | |||
|
Domain Architecture |

|
|||||
| Description | RIM-binding protein 2 (RIM-BP2). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.006779 (rank : 83) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TRI26_HUMAN
|
||||||
| NC score | 0.003537 (rank : 84) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12899 | Gene names | TRIM26, RNF95, ZNF173 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 26 (Zinc finger protein 173) (Acid finger protein) (AFP) (RING finger protein 95). | |||||
|
TCF8_HUMAN
|
||||||
| NC score | 0.002168 (rank : 85) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1057 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P37275, Q12924, Q13800 | Gene names | TCF8, AREB6 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 8 (NIL-2-A zinc finger protein) (Negative regulator of IL2). | |||||
|
AIOL_HUMAN
|
||||||
| NC score | 0.000678 (rank : 86) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UKT9, Q8WWQ9, Q8WWR0, Q8WWR1, Q8WWR2, Q8WWR3 | Gene names | IKZF3, ZNFN1A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein Aiolos (IKAROS family zinc finger protein 3). | |||||
|
ERBB2_MOUSE
|
||||||
| NC score | -0.000177 (rank : 87) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70424, Q61525, Q6ZPE0 | Gene names | Erbb2, Kiaa3023, Neu | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene). | |||||
|
ROR1_MOUSE
|
||||||
| NC score | -0.002027 (rank : 88) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 82 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z139 | Gene names | Ror1, Ntrkr1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase transmembrane receptor ROR1 precursor (EC 2.7.10.1) (Neurotrophic tyrosine kinase, receptor-related 1) (mROR1). | |||||