Please be patient as the page loads
|
ARMX5_MOUSE
|
||||||
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ARMX5_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX5_HUMAN
|
||||||
| θ value | 6.34118e-171 (rank : 2) | NC score | 0.981838 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1M9, Q68DB4, Q9BVZ3, Q9H969 | Gene names | ARMCX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 3.85808e-43 (rank : 3) | NC score | 0.836106 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP1_MOUSE
|
||||||
| θ value | 3.99248e-40 (rank : 4) | NC score | 0.806057 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
GASP2_MOUSE
|
||||||
| θ value | 3.37985e-39 (rank : 5) | NC score | 0.807353 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP2_HUMAN
|
||||||
| θ value | 3.61944e-33 (rank : 6) | NC score | 0.817980 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
ARMX1_HUMAN
|
||||||
| θ value | 5.22648e-32 (rank : 7) | NC score | 0.836695 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
ARMX1_MOUSE
|
||||||
| θ value | 8.3442e-30 (rank : 8) | NC score | 0.831889 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX3_HUMAN
|
||||||
| θ value | 7.06379e-29 (rank : 9) | NC score | 0.818695 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX3_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 10) | NC score | 0.818656 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
ARMX2_HUMAN
|
||||||
| θ value | 3.75424e-22 (rank : 11) | NC score | 0.765109 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| θ value | 4.9032e-22 (rank : 12) | NC score | 0.722854 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ARMX6_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 13) | NC score | 0.603311 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7L4S7, Q9NWJ3 | Gene names | ARMCX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX6_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 14) | NC score | 0.558274 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K3A6, Q3TUH4 | Gene names | Armcx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 15) | NC score | 0.066940 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 16) | NC score | 0.058360 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZC11A_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 17) | NC score | 0.060688 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
HNRPM_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.041833 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
BCLF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.059621 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BCLF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 20) | NC score | 0.054023 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
HNRPM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.036903 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.031099 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.022638 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.036118 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.043460 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.031439 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
CF081_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.181677 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
KCNH5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.014391 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.007939 (rank : 49) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.049085 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CSPG4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.013485 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UVK1, Q92675 | Gene names | CSPG4, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (Melanoma chondroitin sulfate proteoglycan) (Melanoma-associated chondroitin sulfate proteoglycan). | |||||
|
PR285_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.016307 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
SYTL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.010679 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
KCNH5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.014736 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.016412 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
IL4RA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.015501 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |
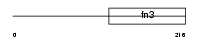
|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.029636 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
JHD2A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.011679 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
RBM28_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.009898 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.001344 (rank : 51) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
BCAS1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.026437 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
H14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.027245 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
NUD12_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.012335 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
SH24A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.011483 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
SMRC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.008986 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
UBE2U_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.006890 (rank : 50) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VVX9, Q8N1D4 | Gene names | UBE2U | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 U (EC 6.3.2.19) (Ubiquitin-protein ligase U) (Ubiquitin carrier protein U). | |||||
|
ARMX4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.407209 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5H9R4, Q5H9K8, Q8N8D6 | Gene names | ARMCX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 4. | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.051912 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF081_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.143489 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.067752 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.050264 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ARMX5_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q3UZB0 | Gene names | Armcx5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX5_HUMAN
|
||||||
| NC score | 0.981838 (rank : 2) | θ value | 6.34118e-171 (rank : 2) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6P1M9, Q68DB4, Q9BVZ3, Q9H969 | Gene names | ARMCX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 5. | |||||
|
ARMX1_HUMAN
|
||||||
| NC score | 0.836695 (rank : 3) | θ value | 5.22648e-32 (rank : 7) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P291, Q53HK2, Q9H2Q0 | Gene names | ARMCX1, ALEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1 (Protein ALEX1) (ARM protein lost in epithelial cancers on chromosome X 1). | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.836106 (rank : 4) | θ value | 3.85808e-43 (rank : 3) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
ARMX1_MOUSE
|
||||||
| NC score | 0.831889 (rank : 5) | θ value | 8.3442e-30 (rank : 8) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9CX83 | Gene names | Armcx1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 1. | |||||
|
ARMX3_HUMAN
|
||||||
| NC score | 0.818695 (rank : 6) | θ value | 7.06379e-29 (rank : 9) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UH62, Q53HC6, Q7LCF5, Q9NPE4 | Gene names | ARMCX3, ALEX3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3 (Protein ALEX3) (ARM protein lost in epithelial cancers on chromosome X 3). | |||||
|
ARMX3_MOUSE
|
||||||
| NC score | 0.818656 (rank : 7) | θ value | 3.50572e-28 (rank : 10) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BHS6, Q91VP8, Q9DC32 | Gene names | Armcx3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 3. | |||||
|
GASP2_HUMAN
|
||||||
| NC score | 0.817980 (rank : 8) | θ value | 3.61944e-33 (rank : 6) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96D09, Q8NAB4 | Gene names | GPRASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP2_MOUSE
|
||||||
| NC score | 0.807353 (rank : 9) | θ value | 3.37985e-39 (rank : 5) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8BUY8 | Gene names | Gprasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 2 (GASP-2). | |||||
|
GASP1_MOUSE
|
||||||
| NC score | 0.806057 (rank : 10) | θ value | 3.99248e-40 (rank : 4) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5U4C1, Q8BKR8, Q8BUN4, Q8BYK9, Q8CHF4, Q8R095 | Gene names | Gprasp1, Kiaa0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
ARMX2_HUMAN
|
||||||
| NC score | 0.765109 (rank : 11) | θ value | 3.75424e-22 (rank : 11) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7L311, O60267, Q5H9D9 | Gene names | ARMCX2, ALEX2, KIAA0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2 (Protein ALEX2) (ARM protein lost in epithelial cancers on chromosome X 2). | |||||
|
ARMX2_MOUSE
|
||||||
| NC score | 0.722854 (rank : 12) | θ value | 4.9032e-22 (rank : 12) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6A058, Q9CXI9 | Gene names | Armcx2, Kiaa0512 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 2. | |||||
|
ARMX6_HUMAN
|
||||||
| NC score | 0.603311 (rank : 13) | θ value | 1.69304e-06 (rank : 13) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7L4S7, Q9NWJ3 | Gene names | ARMCX6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX6_MOUSE
|
||||||
| NC score | 0.558274 (rank : 14) | θ value | 5.44631e-05 (rank : 14) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K3A6, Q3TUH4 | Gene names | Armcx6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ARMCX6. | |||||
|
ARMX4_HUMAN
|
||||||
| NC score | 0.407209 (rank : 15) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q5H9R4, Q5H9K8, Q8N8D6 | Gene names | ARMCX4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Armadillo repeat-containing X-linked protein 4. | |||||
|
CF081_HUMAN
|
||||||
| NC score | 0.181677 (rank : 16) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q5T9G4, Q8NEB2, Q96LL8 | Gene names | C6orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81. | |||||
|
CF081_MOUSE
|
||||||
| NC score | 0.143489 (rank : 17) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80X86, Q9D564, Q9DAN7 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf81 homolog. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.067752 (rank : 18) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.066940 (rank : 19) | θ value | 0.00035302 (rank : 15) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ZC11A_MOUSE
|
||||||
| NC score | 0.060688 (rank : 20) | θ value | 0.0330416 (rank : 17) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6NZF1, Q6NXW9, Q6PDR6, Q80TU7, Q8C5L5, Q99JN6 | Gene names | Zc3h11a, Kiaa0663 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 11A. | |||||
|
BCLF1_HUMAN
|
||||||
| NC score | 0.059621 (rank : 21) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NYF8, Q14673, Q86WU6, Q86WY0 | Gene names | BCLAF1, BTF, KIAA0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.058360 (rank : 22) | θ value | 0.00509761 (rank : 16) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BCLF1_MOUSE
|
||||||
| NC score | 0.054023 (rank : 23) | θ value | 0.47712 (rank : 20) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K019, Q8BNZ0, Q8C2E9, Q9CSW5 | Gene names | Bclaf1, Btf, Kiaa0164 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-associated transcription factor 1 (Btf). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.051912 (rank : 24) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.050264 (rank : 25) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.049085 (rank : 26) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.043460 (rank : 27) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
HNRPM_MOUSE
|
||||||
| NC score | 0.041833 (rank : 28) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D0E1, Q6P1B2, Q99JQ0 | Gene names | Hnrpm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
HNRPM_HUMAN
|
||||||
| NC score | 0.036903 (rank : 29) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52272, Q15584, Q8WZ44, Q96H56, Q9BWL9, Q9Y492 | Gene names | HNRPM | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein M (hnRNP M). | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.036118 (rank : 30) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.031439 (rank : 31) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.031099 (rank : 32) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.029636 (rank : 33) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.027245 (rank : 34) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
BCAS1_MOUSE
|
||||||
| NC score | 0.026437 (rank : 35) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80YN3, Q9CVA1 | Gene names | Bcas1, Nabc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Breast carcinoma amplified sequence 1 homolog (Novel amplified in breast cancer 1 homolog). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.022638 (rank : 36) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.016412 (rank : 37) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.016307 (rank : 38) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
IL4RA_HUMAN
|
||||||
| NC score | 0.015501 (rank : 39) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24394, Q96P01, Q9H181, Q9H182, Q9H183, Q9H184, Q9H185, Q9H186, Q9H187, Q9H188 | Gene names | IL4R, 582J2.1, IL4RA | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (sIL4Ralpha/prot) (IL-4-binding protein) (IL4-BP)]. | |||||
|
KCNH5_HUMAN
|
||||||
| NC score | 0.014736 (rank : 40) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NCM2 | Gene names | KCNH5, EAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (hEAG2). | |||||
|
KCNH5_MOUSE
|
||||||
| NC score | 0.014391 (rank : 41) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q920E3, Q8C035 | Gene names | Kcnh5, Eag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium voltage-gated channel subfamily H member 5 (Voltage-gated potassium channel subunit Kv10.2) (Ether-a-go-go potassium channel 2) (Eag2). | |||||
|
CSPG4_HUMAN
|
||||||
| NC score | 0.013485 (rank : 42) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6UVK1, Q92675 | Gene names | CSPG4, MCSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 4 precursor (Chondroitin sulfate proteoglycan NG2) (Melanoma chondroitin sulfate proteoglycan) (Melanoma-associated chondroitin sulfate proteoglycan). | |||||
|
NUD12_MOUSE
|
||||||
| NC score | 0.012335 (rank : 43) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
JHD2A_MOUSE
|
||||||
| NC score | 0.011679 (rank : 44) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6PCM1, Q2MJQ6, Q3TKW8, Q3UML3, Q6ZQ57, Q8K2J6, Q8K2K4, Q8R350 | Gene names | Jmjd1a, Jhdm2a, Kiaa0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
SH24A_MOUSE
|
||||||
| NC score | 0.011483 (rank : 45) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
SYTL3_MOUSE
|
||||||
| NC score | 0.010679 (rank : 46) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99N48, Q8C506, Q99N47, Q99N49, Q99N54, Q99N79 | Gene names | Sytl3, Slp3 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 3 (Exophilin-6). | |||||
|
RBM28_MOUSE
|
||||||
| NC score | 0.009898 (rank : 47) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CGC6, Q8BG25, Q8VEJ8, Q9CS22, Q9CSE6 | Gene names | Rbm28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 28 (RNA-binding motif protein 28). | |||||
|
SMRC1_HUMAN
|
||||||
| NC score | 0.008986 (rank : 48) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92922, Q6P172, Q8IWH2 | Gene names | SMARCC1, BAF155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.007939 (rank : 49) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
UBE2U_HUMAN
|
||||||
| NC score | 0.006890 (rank : 50) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5VVX9, Q8N1D4 | Gene names | UBE2U | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 U (EC 6.3.2.19) (Ubiquitin-protein ligase U) (Ubiquitin carrier protein U). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.001344 (rank : 51) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 46 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||