Please be patient as the page loads
|
DGLB_HUMAN
|
||||||
| SwissProt Accessions | Q8NCG7, Q6PIX3, Q8N2N2, Q8N9S1, Q8TED3, Q8WXE6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta) (KCCR13L). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DGLB_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NCG7, Q6PIX3, Q8N2N2, Q8N9S1, Q8TED3, Q8WXE6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta) (KCCR13L). | |||||
|
DGLB_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998044 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WC9, Q8BU39, Q8BU97, Q8BV05 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta). | |||||
|
DGLA_MOUSE
|
||||||
| θ value | 2.62879e-100 (rank : 3) | NC score | 0.910628 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WQJ1, Q6ZQ76 | Gene names | Nsddr, Kiaa0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
DGLA_HUMAN
|
||||||
| θ value | 3.79596e-99 (rank : 4) | NC score | 0.910956 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4D2, Q6WQJ0 | Gene names | NSDDR, C11orf11, KIAA0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
GASP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.028111 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
BAZ2A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 6) | NC score | 0.008940 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
LRP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 7) | NC score | 0.008364 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |
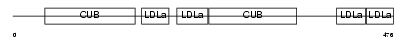
|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
LRP5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.006028 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
APTX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.012878 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.002163 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
NCOA3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.007424 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
SIX4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.005864 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIU6 | Gene names | SIX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX4 (Sine oculis homeobox homolog 4). | |||||
|
DGLB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8NCG7, Q6PIX3, Q8N2N2, Q8N9S1, Q8TED3, Q8WXE6 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta) (KCCR13L). | |||||
|
DGLB_MOUSE
|
||||||
| NC score | 0.998044 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WC9, Q8BU39, Q8BU97, Q8BV05 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase beta (EC 3.1.1.-) (DGL-beta). | |||||
|
DGLA_HUMAN
|
||||||
| NC score | 0.910956 (rank : 3) | θ value | 3.79596e-99 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4D2, Q6WQJ0 | Gene names | NSDDR, C11orf11, KIAA0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
DGLA_MOUSE
|
||||||
| NC score | 0.910628 (rank : 4) | θ value | 2.62879e-100 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6WQJ1, Q6ZQ76 | Gene names | Nsddr, Kiaa0659 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sn1-specific diacylglycerol lipase alpha (EC 3.1.1.-) (DGL-alpha) (Neural stem cell-derived dendrite regulator). | |||||
|
GASP1_HUMAN
|
||||||
| NC score | 0.028111 (rank : 5) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5JY77, O43168, Q96LA1 | Gene names | GPRASP1, GASP, KIAA0443 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor-associated sorting protein 1 (GASP-1). | |||||
|
APTX_HUMAN
|
||||||
| NC score | 0.012878 (rank : 6) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
BAZ2A_MOUSE
|
||||||
| NC score | 0.008940 (rank : 7) | θ value | 5.27518 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91YE5 | Gene names | Baz2a, Tip5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5). | |||||
|
LRP3_HUMAN
|
||||||
| NC score | 0.008364 (rank : 8) | θ value | 6.88961 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75074 | Gene names | LRP3 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 3 precursor (hLRp105). | |||||
|
NCOA3_MOUSE
|
||||||
| NC score | 0.007424 (rank : 9) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O09000, Q9CRD5 | Gene names | Ncoa3, Aib1, Pcip, Rac3, Tram1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor coactivator 3 (EC 2.3.1.48) (NCoA-3) (Thyroid hormone receptor activator molecule 1) (TRAM-1) (ACTR) (Receptor-associated coactivator 3) (RAC-3) (Amplified in breast cancer-1 protein homolog) (AIB-1) (Steroid receptor coactivator protein 3) (SRC-3) (CBP- interacting protein) (p/CIP) (pCIP). | |||||
|
LRP5_MOUSE
|
||||||
| NC score | 0.006028 (rank : 10) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VN0, O88883, Q9R208 | Gene names | Lrp5, Lrp7 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 5 precursor (LRP7) (Lr3). | |||||
|
SIX4_HUMAN
|
||||||
| NC score | 0.005864 (rank : 11) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UIU6 | Gene names | SIX4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein SIX4 (Sine oculis homeobox homolog 4). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.002163 (rank : 12) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||