Please be patient as the page loads
|
ANC1_MOUSE
|
||||||
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ANC1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993552 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1A4, Q9BSE6, Q9H8D0 | Gene names | ANAPC1, TSG24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
ANC1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
COE1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 3) | NC score | 0.066641 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 4) | NC score | 0.066641 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 0.279714 (rank : 5) | NC score | 0.031945 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FBX38_MOUSE
|
||||||
| θ value | 0.365318 (rank : 6) | NC score | 0.049961 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
YIPF5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.072838 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQQ2 | Gene names | Yipf5, Yip1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF5 (YIP1 family member 5) (YPT-interacting protein 1 A). | |||||
|
RBP26_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.039533 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
FIGN_MOUSE
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.028995 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.024699 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
YAP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.050745 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
KCNT1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.041494 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.052950 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
JHD3B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.014604 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
KCNT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.038478 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
PSMD2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.050643 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
PSMD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.050081 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.024439 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
GRIP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.012427 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
YAP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.037165 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
COE3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.051184 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.051221 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
LV1A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.009776 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01699 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda chain V-I region VOR. | |||||
|
RGS14_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.012452 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.011214 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.053616 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PMS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.029523 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |
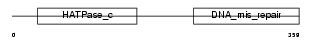
|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
REPS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.023968 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |
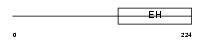
|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
SMD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.053730 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62314, P13641 | Gene names | SNRPD1 | |||
|
Domain Architecture |
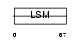
|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
SMD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.053730 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62315, P13641, Q3UYM0 | Gene names | Snrpd1 | |||
|
Domain Architecture |
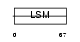
|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
YIPF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.040903 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969M3, Q4VSN6, Q53EX4, Q8NHE5, Q9H338, Q9H3U4 | Gene names | YIPF5, FINGER5, YIP1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF5 (YIP1 family member 5) (YPT-interacting protein 1 A) (Five-pass transmembrane protein localizing in the Golgi apparatus and the endoplasmic reticulum 5) (Smooth muscle cell-associated protein 5) (SMAP-5). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.012042 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BNC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.015867 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.015771 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
EPIPL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.010649 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
FBX38_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.035236 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.016144 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
STCH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.014410 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
GPR92_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.001802 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1C0 | Gene names | GPR92, GPR93 | |||
|
Domain Architecture |
|
|||||
| Description | Probable G-protein coupled receptor 92. | |||||
|
KLF5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.000794 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.012795 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
TEX15_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.021697 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
ANC1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P53995, Q8BP33, Q8C772 | Gene names | Anapc1, Mcpr, Tsg24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
ANC1_HUMAN
|
||||||
| NC score | 0.993552 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9H1A4, Q9BSE6, Q9H8D0 | Gene names | ANAPC1, TSG24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Anaphase-promoting complex subunit 1 (APC1) (Cyclosome subunit 1) (Protein Tsg24) (Mitotic checkpoint regulator). | |||||
|
YIPF5_MOUSE
|
||||||
| NC score | 0.072838 (rank : 3) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQQ2 | Gene names | Yipf5, Yip1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF5 (YIP1 family member 5) (YPT-interacting protein 1 A). | |||||
|
COE1_HUMAN
|
||||||
| NC score | 0.066641 (rank : 4) | θ value | 0.163984 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UH73, Q8IW11 | Gene names | EBF1, COE1, EBF | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
COE1_MOUSE
|
||||||
| NC score | 0.066641 (rank : 5) | θ value | 0.163984 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q07802 | Gene names | Ebf1, Coe1, Ebf | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE1 (OE-1) (O/E-1) (Early B-cell factor). | |||||
|
SMD1_HUMAN
|
||||||
| NC score | 0.053730 (rank : 6) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62314, P13641 | Gene names | SNRPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
SMD1_MOUSE
|
||||||
| NC score | 0.053730 (rank : 7) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P62315, P13641, Q3UYM0 | Gene names | Snrpd1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein Sm D1 (snRNP core protein D1) (Sm-D1) (Sm-D autoantigen). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.053616 (rank : 8) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.052950 (rank : 9) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
COE3_MOUSE
|
||||||
| NC score | 0.051221 (rank : 10) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08791, O08793 | Gene names | Ebf3, Coe3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
COE3_HUMAN
|
||||||
| NC score | 0.051184 (rank : 11) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4W6, Q5T6H9, Q9H4W5 | Gene names | EBF3, COE3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor COE3 (Early B-cell factor 3) (EBF-3) (Olf-1/EBF- like 2) (OE-2) (O/E-2). | |||||
|
YAP1_HUMAN
|
||||||
| NC score | 0.050745 (rank : 12) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46937 | Gene names | YAP1, YAP65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
PSMD2_HUMAN
|
||||||
| NC score | 0.050643 (rank : 13) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13200, Q12932, Q15321, Q96I12 | Gene names | PSMD2, TRAP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97) (Tumor necrosis factor type 1 receptor- associated protein 2) (55.11 protein). | |||||
|
PSMD2_MOUSE
|
||||||
| NC score | 0.050081 (rank : 14) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDM4 | Gene names | Psmd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 2 (26S proteasome regulatory subunit RPN1) (26S proteasome regulatory subunit S2) (26S proteasome subunit p97). | |||||
|
FBX38_MOUSE
|
||||||
| NC score | 0.049961 (rank : 15) | θ value | 0.365318 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMI0, Q8BTQ4, Q8C0H4 | Gene names | Fbxo38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity) (MoKA). | |||||
|
KCNT1_HUMAN
|
||||||
| NC score | 0.041494 (rank : 16) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JUK3, Q9P2C5 | Gene names | KCNT1, KIAA1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1 (KCa4.1). | |||||
|
YIPF5_HUMAN
|
||||||
| NC score | 0.040903 (rank : 17) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q969M3, Q4VSN6, Q53EX4, Q8NHE5, Q9H338, Q9H3U4 | Gene names | YIPF5, FINGER5, YIP1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein YIPF5 (YIP1 family member 5) (YPT-interacting protein 1 A) (Five-pass transmembrane protein localizing in the Golgi apparatus and the endoplasmic reticulum 5) (Smooth muscle cell-associated protein 5) (SMAP-5). | |||||
|
RBP26_HUMAN
|
||||||
| NC score | 0.039533 (rank : 18) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q68DN6, Q6V1X0 | Gene names | RANBP2L6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 6 (RanBP2L6). | |||||
|
KCNT1_MOUSE
|
||||||
| NC score | 0.038478 (rank : 19) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPR4, Q8C3E7 | Gene names | Kcnt1, Kiaa1422 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Potassium channel subfamily T member 1. | |||||
|
YAP1_MOUSE
|
||||||
| NC score | 0.037165 (rank : 20) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46938, Q52KJ5 | Gene names | Yap1, Yap, Yap65 | |||
|
Domain Architecture |

|
|||||
| Description | 65 kDa Yes-associated protein (YAP65). | |||||
|
FBX38_HUMAN
|
||||||
| NC score | 0.035236 (rank : 21) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PIJ6, Q6PK72, Q7Z2U0, Q86VN3, Q9BXY6, Q9H837, Q9HC40 | Gene names | FBXO38 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | F-box only protein 38 (Modulator of KLF7 activity homolog) (MoKA). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.031945 (rank : 22) | θ value | 0.279714 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PMS2_MOUSE
|
||||||
| NC score | 0.029523 (rank : 23) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54279 | Gene names | Pms2 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 2 (DNA mismatch repair protein PMS2). | |||||
|
FIGN_MOUSE
|
||||||
| NC score | 0.028995 (rank : 24) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERZ6, Q3TPB0, Q3UP57, Q6PCM0 | Gene names | Fign | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.024699 (rank : 25) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.024439 (rank : 26) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
REPS2_MOUSE
|
||||||
| NC score | 0.023968 (rank : 27) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
TEX15_HUMAN
|
||||||
| NC score | 0.021697 (rank : 28) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BXT5 | Gene names | TEX15 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 15 protein. | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.016144 (rank : 29) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
BNC2_HUMAN
|
||||||
| NC score | 0.015867 (rank : 30) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6ZN30, Q6T3A3, Q8NAR2, Q9H6J0, Q9NXV0 | Gene names | BNC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
BNC2_MOUSE
|
||||||
| NC score | 0.015771 (rank : 31) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMQ3, Q6T3A4 | Gene names | Bnc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein basonuclin-2. | |||||
|
JHD3B_MOUSE
|
||||||
| NC score | 0.014604 (rank : 32) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VY5, Q3UR22, Q6ZQ30, Q99K42 | Gene names | Jmjd2b, Jhdm3b | |||
|
Domain Architecture |

|
|||||
| Description | JmjC domain-containing histone demethylation protein 3B (EC 1.14.11.-) (Jumonji domain-containing protein 2B). | |||||
|
STCH_HUMAN
|
||||||
| NC score | 0.014410 (rank : 33) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48723, Q8NE40 | Gene names | STCH | |||
|
Domain Architecture |

|
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.012795 (rank : 34) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
RGS14_MOUSE
|
||||||
| NC score | 0.012452 (rank : 35) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97492, Q9DCD1 | Gene names | Rgs14 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 14 (RGS14) (RAP1/RAP2-interacting protein). | |||||
|
GRIP1_HUMAN
|
||||||
| NC score | 0.012427 (rank : 36) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3R0 | Gene names | GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutamate receptor-interacting protein 1 (GRIP1 protein). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.012042 (rank : 37) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.011214 (rank : 38) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
EPIPL_HUMAN
|
||||||
| NC score | 0.010649 (rank : 39) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P58107, Q76E58 | Gene names | EPPK1, EPIPL | |||
|
Domain Architecture |

|
|||||
| Description | Epiplakin (450 kDa epidermal antigen). | |||||
|
LV1A_HUMAN
|
||||||
| NC score | 0.009776 (rank : 40) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P01699 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig lambda chain V-I region VOR. | |||||
|
GPR92_HUMAN
|
||||||
| NC score | 0.001802 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1C0 | Gene names | GPR92, GPR93 | |||
|
Domain Architecture |

|
|||||
| Description | Probable G-protein coupled receptor 92. | |||||
|
KLF5_HUMAN
|
||||||
| NC score | 0.000794 (rank : 42) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||