Please be patient as the page loads
|
MA1A2_MOUSE
|
||||||
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MA1A1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992370 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33908, Q9NU44, Q9UJI3 | Gene names | MAN1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.991276 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998265 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60476, Q9H510 | Gene names | MAN1A2, MAN1B | |||
|
Domain Architecture |
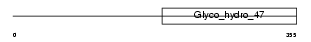
|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1C1_HUMAN
|
||||||
| θ value | 1.19867e-161 (rank : 5) | NC score | 0.988042 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
MA1B1_HUMAN
|
||||||
| θ value | 2.30828e-88 (rank : 6) | NC score | 0.930026 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
EDEM2_HUMAN
|
||||||
| θ value | 4.71619e-41 (rank : 7) | NC score | 0.871812 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV94, Q6IA89, Q6UWZ4, Q9H4U0, Q9H886, Q9NTL9, Q9NVE6 | Gene names | EDEM2, C20orf31, C20orf49 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 2 precursor. | |||||
|
EDEM1_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 8) | NC score | 0.835210 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM1_MOUSE
|
||||||
| θ value | 6.15952e-41 (rank : 9) | NC score | 0.834665 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM3_HUMAN
|
||||||
| θ value | 9.20422e-37 (rank : 10) | NC score | 0.821882 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.019365 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.012537 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.020559 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
NF2L3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.014470 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.012364 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.023350 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.008544 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.005482 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.006765 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RNF32_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.015324 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
IFT74_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.014891 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.009237 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.007653 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.007747 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
INVS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.002157 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
CACB4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.008783 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00305, O60515, Q96L40 | Gene names | CACNB4, CACNLB4 | |||
|
Domain Architecture |
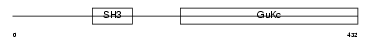
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.008873 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0S4, Q8BRN6, Q8CAJ9 | Gene names | Cacnb4, Cacnlb4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
HOME2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.006937 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
REPS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.008538 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.004754 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BTBD7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.010226 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
MA1A2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P39098, Q5SUY6, Q5SUY7, Q60599, Q9CUY9 | Gene names | Man1a2, Man1b | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A2_HUMAN
|
||||||
| NC score | 0.998265 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60476, Q9H510 | Gene names | MAN1A2, MAN1B | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IB) (Alpha-1,2-mannosidase IB) (Mannosidase alpha class 1A member 2). | |||||
|
MA1A1_HUMAN
|
||||||
| NC score | 0.992370 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P33908, Q9NU44, Q9UJI3 | Gene names | MAN1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1A1_MOUSE
|
||||||
| NC score | 0.991276 (rank : 4) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P45700 | Gene names | Man1a1, Man1a | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IA) (Alpha-1,2-mannosidase IA) (Mannosidase alpha class 1A member 1) (Man(9)-alpha-mannosidase) (Man9-mannosidase). | |||||
|
MA1C1_HUMAN
|
||||||
| NC score | 0.988042 (rank : 5) | θ value | 1.19867e-161 (rank : 5) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9NR34, Q9Y545 | Gene names | MAN1C1, MAN1A3, MAN1C | |||
|
Domain Architecture |

|
|||||
| Description | Mannosyl-oligosaccharide 1,2-alpha-mannosidase IC (EC 3.2.1.113) (Processing alpha-1,2-mannosidase IC) (Alpha-1,2-mannosidase IC) (Mannosidase alpha class 1C member 1) (HMIC). | |||||
|
MA1B1_HUMAN
|
||||||
| NC score | 0.930026 (rank : 6) | θ value | 2.30828e-88 (rank : 6) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UKM7, Q5VSG3, Q9BRS9, Q9Y5K7 | Gene names | MAN1B1 | |||
|
Domain Architecture |

|
|||||
| Description | Endoplasmic reticulum mannosyl-oligosaccharide 1,2-alpha-mannosidase (EC 3.2.1.113) (ER alpha-1,2-mannosidase) (Mannosidase alpha class 1B member 1) (Man9GlcNAc2-specific-processing alpha-mannosidase). | |||||
|
EDEM2_HUMAN
|
||||||
| NC score | 0.871812 (rank : 7) | θ value | 4.71619e-41 (rank : 7) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BV94, Q6IA89, Q6UWZ4, Q9H4U0, Q9H886, Q9NTL9, Q9NVE6 | Gene names | EDEM2, C20orf31, C20orf49 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 2 precursor. | |||||
|
EDEM1_HUMAN
|
||||||
| NC score | 0.835210 (rank : 8) | θ value | 6.15952e-41 (rank : 8) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92611 | Gene names | EDEM1, EDEM, KIAA0212 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM1_MOUSE
|
||||||
| NC score | 0.834665 (rank : 9) | θ value | 6.15952e-41 (rank : 9) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925U4 | Gene names | Edem1, Edem | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 1. | |||||
|
EDEM3_HUMAN
|
||||||
| NC score | 0.821882 (rank : 10) | θ value | 9.20422e-37 (rank : 10) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BZQ6, Q5TEZ0, Q9HCW1, Q9UFV7 | Gene names | EDEM3, C1orf22 | |||
|
Domain Architecture |

|
|||||
| Description | ER degradation-enhancing alpha-mannosidase-like 3. | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.023350 (rank : 11) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.020559 (rank : 12) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.019365 (rank : 13) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
RNF32_HUMAN
|
||||||
| NC score | 0.015324 (rank : 14) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.014891 (rank : 15) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
NF2L3_HUMAN
|
||||||
| NC score | 0.014470 (rank : 16) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y4A8, Q6NUS0, Q7Z498, Q86UJ4, Q86VR5, Q9UQA4 | Gene names | NFE2L3, NRF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor erythroid 2-related factor 3 (NF-E2-related factor 3) (NFE2-related factor 3) (Nuclear factor, erythroid derived 2, like 3). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.012537 (rank : 17) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.012364 (rank : 18) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
BTBD7_MOUSE
|
||||||
| NC score | 0.010226 (rank : 19) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CFE5, Q80TC3, Q8BZY9 | Gene names | Btbd7, Kiaa1525 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 7. | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.009237 (rank : 20) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
CACB4_MOUSE
|
||||||
| NC score | 0.008873 (rank : 21) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R0S4, Q8BRN6, Q8CAJ9 | Gene names | Cacnb4, Cacnlb4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CACB4_HUMAN
|
||||||
| NC score | 0.008783 (rank : 22) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00305, O60515, Q96L40 | Gene names | CACNB4, CACNLB4 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-4 (CAB4) (Calcium channel voltage-dependent subunit beta 4). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.008544 (rank : 23) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
REPS1_HUMAN
|
||||||
| NC score | 0.008538 (rank : 24) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96D71, Q8NDR7, Q8WU62, Q9BXY9 | Gene names | REPS1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 1 (RalBP1-interacting protein 1). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.007747 (rank : 25) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.007653 (rank : 26) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
HOME2_MOUSE
|
||||||
| NC score | 0.006937 (rank : 27) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QWW1, O89025, Q9Z0E4 | Gene names | Homer2, Vesl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 2 (Homer-2) (Cupidin) (VASP/Ena-related gene up- regulated during seizure and LTP 2) (Vesl-2). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.006765 (rank : 28) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.005482 (rank : 29) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.004754 (rank : 30) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.002157 (rank : 31) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 31 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||