Please be patient as the page loads
|
SAPS3_MOUSE
|
||||||
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.820220 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.958997 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.972495 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.971248 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS3_HUMAN
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992485 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
SAPS3_MOUSE
|
||||||
| θ value | 0 (rank : 6) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 7) | NC score | 0.032669 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 8) | NC score | 0.032412 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.026446 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
CN145_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.021750 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
NOP14_MOUSE
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.035131 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.037141 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.026821 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.025588 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.025201 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.020536 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.020450 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.054048 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
GRIN1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.048290 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.018208 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.051791 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
AP2A2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.028995 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
AP2A2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.029237 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
NIPA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.025985 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
SIN3B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.017480 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |
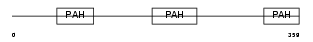
|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
CASC3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.034882 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
NF2L2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.018379 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
RAE2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.024364 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.014897 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
TOP2A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.018432 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
ADDA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.027841 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
ADDB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.020772 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.033017 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.042691 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MYST3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.021017 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.033781 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
SUHW4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.004692 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
TP53B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.030215 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
TP53B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.056558 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
ANR26_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.007708 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.035084 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
GP1BA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.012373 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.016275 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
MYBPH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.012881 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |
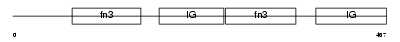
|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
SYTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.009153 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.014037 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.025620 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.037623 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CCNB3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.011773 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
GTPB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.024678 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
GTPB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.024684 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
JHD2C_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.018609 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
OSBP1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.013793 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
RUSC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.012120 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
UBQL4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.015784 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRR5, Q9BR98, Q9UHX4 | Gene names | UBQLN4, C1orf6, UBIN | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.033693 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
CNTN6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.003244 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
CNTN6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.003175 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||
|
DNL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.014534 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.012770 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
LHX2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.003247 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |
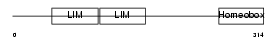
|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.036837 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.008124 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.007934 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.035601 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
WDR7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.011435 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E6, Q86UX5, Q86VP2, Q96PS7 | Gene names | WDR7, KIAA0541, TRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 7 (TGF-beta resistance-associated protein TRAG) (Rabconnectin-3 beta). | |||||
|
E41L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.006142 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
EDD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.011722 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
HCN3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.003702 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.014586 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
LPIN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.011653 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.010109 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NOP56_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.008406 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
NP1L3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.011232 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
PVRL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.008654 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32507, Q62096 | Gene names | Pvrl2, Mph, Pvr, Pvs | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 2 precursor (Murine herpesvirus entry protein B) (mHveB) (Nectin-2) (Poliovirus receptor homolog) (CD112 antigen). | |||||
|
R3HD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.018918 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
T240L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.008280 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.068615 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
SAPS3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | Q922D4, Q6ZPM9, Q8BTW6, Q9D2X9 | Gene names | Saps3, D19Ertd703e, Kiaa1558 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3. | |||||
|
SAPS3_HUMAN
|
||||||
| NC score | 0.992485 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q5H9R7, Q3B7I1, Q3I4Y0, Q3KR35, Q68CR3, Q7L4R8, Q8N3B2, Q96MB2, Q9H2K5, Q9H2K6, Q9HCL4, Q9NUY3 | Gene names | SAPS3, C11orf23, KIAA1558, PP6R3, SAPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 3 (Sporulation-induced transcript 4- associated protein SAPL) (Protein phosphatase 6 regulatory subunit 3). | |||||
|
SAPS2_HUMAN
|
||||||
| NC score | 0.972495 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS2_MOUSE
|
||||||
| NC score | 0.971248 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R3Q2, Q9CXB7 | Gene names | Saps2, Kiaa0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
SAPS1_MOUSE
|
||||||
| NC score | 0.958997 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7TSI3, Q80TJ4 | Gene names | Saps1, Kiaa1115 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SAPS domain family member 1. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.820220 (rank : 6) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.068615 (rank : 7) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
TP53B_MOUSE
|
||||||
| NC score | 0.056558 (rank : 8) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P70399, Q68FD0, Q8CI97, Q91YC9 | Gene names | Tp53bp1, Trp53bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.054048 (rank : 9) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.051791 (rank : 10) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GRIN1_MOUSE
|
||||||
| NC score | 0.048290 (rank : 11) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q3UNH4, Q6PGF9, Q9QZY2 | Gene names | Gprin1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein-regulated inducer of neurite outgrowth 1 (GRIN1). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.042691 (rank : 12) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.037623 (rank : 13) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.037141 (rank : 14) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.036837 (rank : 15) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.035601 (rank : 16) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
NOP14_MOUSE
|
||||||
| NC score | 0.035131 (rank : 17) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8R3N1 | Gene names | Nop14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable nucleolar complex protein 14. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.035084 (rank : 18) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CASC3_MOUSE
|
||||||
| NC score | 0.034882 (rank : 19) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8K3W3, Q8K219, Q99NF0 | Gene names | Casc3, Mln51 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC3 (Cancer susceptibility candidate gene 3 protein homolog) (Metastatic lymph node protein 51 homolog) (Protein MLN 51 homolog) (Protein barentsz) (Btz). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.033781 (rank : 20) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.033693 (rank : 21) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.033017 (rank : 22) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.032669 (rank : 23) | θ value | 0.0113563 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.032412 (rank : 24) | θ value | 0.0330416 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
TP53B_HUMAN
|
||||||
| NC score | 0.030215 (rank : 25) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q12888, Q2M1Z7, Q4LE46, Q5FWZ3, Q7Z3U4 | Gene names | TP53BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor suppressor p53-binding protein 1 (p53-binding protein 1) (p53BP1) (53BP1). | |||||
|
AP2A2_MOUSE
|
||||||
| NC score | 0.029237 (rank : 26) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P17427, Q921V0 | Gene names | Ap2a2, Adtab | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit). | |||||
|
AP2A2_HUMAN
|
||||||
| NC score | 0.028995 (rank : 27) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94973, O75403, Q96SI8 | Gene names | AP2A2, ADTAB, KIAA0899 | |||
|
Domain Architecture |

|
|||||
| Description | AP-2 complex subunit alpha-2 (Adapter-related protein complex 2 alpha- 2 subunit) (Alpha-adaptin C) (Adaptor protein complex AP-2 alpha-2 subunit) (Clathrin assembly protein complex 2 alpha-C large chain) (100 kDa coated vesicle protein C) (Plasma membrane adaptor HA2/AP2 adaptin alpha C subunit) (Huntingtin-interacting protein HYPJ). | |||||
|
ADDA_MOUSE
|
||||||
| NC score | 0.027841 (rank : 28) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QYC0, Q9JLE3 | Gene names | Add1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-adducin (Erythrocyte adducin subunit alpha). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.026821 (rank : 29) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.026446 (rank : 30) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
NIPA_MOUSE
|
||||||
| NC score | 0.025985 (rank : 31) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80YV2, Q3TJE6, Q80Z11, Q8BTW5, Q8CI56, Q8R3U7 | Gene names | Zc3hc1, Nipa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear-interacting partner of ALK (Nuclear-interacting partner of anaplastic lymphoma kinase) (mNIPA) (Zinc finger C3HC-type protein 1). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.025620 (rank : 32) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.025588 (rank : 33) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.025201 (rank : 34) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
GTPB2_MOUSE
|
||||||
| NC score | 0.024684 (rank : 35) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UJK4, Q9EST7, Q9JIX7 | Gene names | Gtpbp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2 (GTP-binding-like protein 2). | |||||
|
GTPB2_HUMAN
|
||||||
| NC score | 0.024678 (rank : 36) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BX10, Q5T7E8, Q8ND84, Q8TAH7, Q8WUA5, Q9HCS9, Q9NRU4, Q9NX60 | Gene names | GTPBP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GTP-binding protein 2. | |||||
|
RAE2_MOUSE
|
||||||
| NC score | 0.024364 (rank : 37) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9QZD5, Q8CH16 | Gene names | Chml, Rep2 | |||
|
Domain Architecture |

|
|||||
| Description | Rab proteins geranylgeranyltransferase component A 2 (Rab escort protein 2) (REP-2) (Choroideraemia-like protein). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.021750 (rank : 38) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
MYST3_HUMAN
|
||||||
| NC score | 0.021017 (rank : 39) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q92794 | Gene names | MYST3, MOZ, RUNXBP2, ZNF220 | |||
|
Domain Architecture |

|
|||||
| Description | Histone acetyltransferase MYST3 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 3) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3) (Runt-related transcription factor-binding protein 2) (Monocytic leukemia zinc finger protein) (Zinc finger protein 220). | |||||
|
ADDB_MOUSE
|
||||||
| NC score | 0.020772 (rank : 40) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 574 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9QYB8, Q3U0E1, Q80VH9, Q8C1C4, Q9CXE3, Q9JLE5, Q9QYB7 | Gene names | Add2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-adducin (Erythrocyte adducin subunit beta) (Add97). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.020536 (rank : 41) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.020450 (rank : 42) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
R3HD1_HUMAN
|
||||||
| NC score | 0.018918 (rank : 43) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q15032, Q8IW32 | Gene names | R3HDM1, KIAA0029, R3HDM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | R3H domain-containing protein 1. | |||||
|
JHD2C_MOUSE
|
||||||
| NC score | 0.018609 (rank : 44) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q69ZK6, Q6NV48, Q8BUF5, Q8C4I5, Q8C5Q9 | Gene names | Jmjd1c, Jhdm2c, Kiaa1380 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable JmjC domain-containing histone demethylation protein 2C (EC 1.14.11.-) (Jumonji domain-containing protein 1C). | |||||
|
TOP2A_MOUSE
|
||||||
| NC score | 0.018432 (rank : 45) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01320 | Gene names | Top2a, Top-2, Top2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA topoisomerase 2-alpha (EC 5.99.1.3) (DNA topoisomerase II, alpha isozyme). | |||||
|
NF2L2_HUMAN
|
||||||
| NC score | 0.018379 (rank : 46) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q16236, Q96F71 | Gene names | NFE2L2, NRF2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor erythroid 2-related factor 2 (NF-E2-related factor 2) (NFE2-related factor 2) (Nuclear factor, erythroid derived 2, like 2) (HEBP1). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.018208 (rank : 47) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
SIN3B_HUMAN
|
||||||
| NC score | 0.017480 (rank : 48) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75182, Q68GC2, Q6P4B8, Q8TB34, Q9BSC8 | Gene names | SIN3B, KIAA0700 | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3b (Transcriptional corepressor Sin3b) (Histone deacetylase complex subunit Sin3b). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.016275 (rank : 49) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
UBQL4_HUMAN
|
||||||
| NC score | 0.015784 (rank : 50) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRR5, Q9BR98, Q9UHX4 | Gene names | UBQLN4, C1orf6, UBIN | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-4 (Ataxin-1 ubiquitin-like-interacting protein A1U). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.014897 (rank : 51) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.014586 (rank : 52) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
DNL1_MOUSE
|
||||||
| NC score | 0.014534 (rank : 53) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P37913 | Gene names | Lig1, Lig-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.014037 (rank : 54) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
OSBP1_HUMAN
|
||||||
| NC score | 0.013793 (rank : 55) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P22059 | Gene names | OSBP, OSBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein 1. | |||||
|
MYBPH_MOUSE
|
||||||
| NC score | 0.012881 (rank : 56) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P70402 | Gene names | Mybph | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-binding protein H (MyBP-H) (H-protein). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.012770 (rank : 57) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
GP1BA_HUMAN
|
||||||
| NC score | 0.012373 (rank : 58) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P07359, Q14441, Q16469, Q8N1F3, Q8NG39, Q9HDC7, Q9UEK1, Q9UQS4 | Gene names | GP1BA | |||
|
Domain Architecture |

|
|||||
| Description | Platelet glycoprotein Ib alpha chain precursor (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (Antigen CD42b-alpha) (CD42b antigen) [Contains: Glycocalicin]. | |||||
|
RUSC1_MOUSE
|
||||||
| NC score | 0.012120 (rank : 59) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BG26, Q6PHT9 | Gene names | Rusc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 1. | |||||
|
CCNB3_HUMAN
|
||||||
| NC score | 0.011773 (rank : 60) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WWL7, Q96SB5, Q96SB6, Q96SB7, Q9NT38 | Gene names | CCNB3, CYCB3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
EDD1_HUMAN
|
||||||
| NC score | 0.011722 (rank : 61) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95071, O94970, Q9NPL3 | Gene names | EDD1, EDD, HYD, KIAA0896 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin-protein ligase EDD1 (EC 6.3.2.-) (Hyperplastic discs protein homolog) (hHYD) (Progestin-induced protein). | |||||
|
LPIN2_HUMAN
|
||||||
| NC score | 0.011653 (rank : 62) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q92539 | Gene names | LPIN2, KIAA0249 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipin-2. | |||||
|
WDR7_HUMAN
|
||||||
| NC score | 0.011435 (rank : 63) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E6, Q86UX5, Q86VP2, Q96PS7 | Gene names | WDR7, KIAA0541, TRAG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 7 (TGF-beta resistance-associated protein TRAG) (Rabconnectin-3 beta). | |||||
|
NP1L3_MOUSE
|
||||||
| NC score | 0.011232 (rank : 64) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q794H2, O54802 | Gene names | Nap1l3, Mb20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome assembly protein 1-like 3 (Brain-specific protein MB20). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.010109 (rank : 65) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
SYTL1_MOUSE
|
||||||
| NC score | 0.009153 (rank : 66) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99N80, Q99J26 | Gene names | Sytl1, Slp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 1 (Exophilin-7). | |||||
|
PVRL2_MOUSE
|
||||||
| NC score | 0.008654 (rank : 67) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32507, Q62096 | Gene names | Pvrl2, Mph, Pvr, Pvs | |||
|
Domain Architecture |

|
|||||
| Description | Poliovirus receptor-related protein 2 precursor (Murine herpesvirus entry protein B) (mHveB) (Nectin-2) (Poliovirus receptor homolog) (CD112 antigen). | |||||
|
NOP56_HUMAN
|
||||||
| NC score | 0.008406 (rank : 68) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
T240L_MOUSE
|
||||||
| NC score | 0.008280 (rank : 69) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6JPI3, Q3TRF4, Q3UQI8, Q80TM3, Q80WQ0 | Gene names | Thrap2, Kiaa1025, Trap240l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 2 (Thyroid hormone receptor-associated protein complex 240 kDa component-like). | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.008124 (rank : 70) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.007934 (rank : 71) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
ANR26_HUMAN
|
||||||
| NC score | 0.007708 (rank : 72) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1614 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UPS8, Q2TAZ3, Q6ZR14, Q9H1Q1, Q9NSK9, Q9NTD5, Q9NW69 | Gene names | ANKRD26, KIAA1074 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 26. | |||||
|
E41L3_MOUSE
|
||||||
| NC score | 0.006142 (rank : 73) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9WV92, Q9R102 | Gene names | Epb41l3, Dal1, Epb4.1l3 | |||
|
Domain Architecture |

|
|||||
| Description | Band 4.1-like protein 3 (4.1B) (Differentially expressed in adenocarcinoma of the lung protein 1) (DAL-1) (DAL1P) (mDAL-1). | |||||
|
SUHW4_MOUSE
|
||||||
| NC score | 0.004692 (rank : 74) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
HCN3_MOUSE
|
||||||
| NC score | 0.003702 (rank : 75) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88705 | Gene names | Hcn3, Hac3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 (Hyperpolarization-activated cation channel 3) (HAC-3). | |||||
|
LHX2_HUMAN
|
||||||
| NC score | 0.003247 (rank : 76) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P50458, O95860, Q8N1Z3 | Gene names | LHX2, LH2 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx2 (Homeobox protein LH-2). | |||||
|
CNTN6_HUMAN
|
||||||
| NC score | 0.003244 (rank : 77) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ52 | Gene names | CNTN6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (hNB-3). | |||||
|
CNTN6_MOUSE
|
||||||
| NC score | 0.003175 (rank : 78) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JMB8, Q8C6X1 | Gene names | Cntn6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Contactin-6 precursor (Neural recognition molecule NB-3) (mNB-3). | |||||