Please be patient as the page loads
|
TM1L1_MOUSE
|
||||||
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TM1L1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992571 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 2.03627e-60 (rank : 3) | NC score | 0.910587 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| θ value | 5.0155e-59 (rank : 4) | NC score | 0.909091 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
GGA2_HUMAN
|
||||||
| θ value | 6.20254e-17 (rank : 5) | NC score | 0.651067 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 2.35696e-16 (rank : 6) | NC score | 0.675196 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_MOUSE
|
||||||
| θ value | 6.85773e-16 (rank : 7) | NC score | 0.654495 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_HUMAN
|
||||||
| θ value | 1.16975e-15 (rank : 8) | NC score | 0.652311 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 9) | NC score | 0.645011 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 3.40345e-15 (rank : 10) | NC score | 0.656398 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 11) | NC score | 0.485835 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM2_MOUSE
|
||||||
| θ value | 1.2105e-12 (rank : 12) | NC score | 0.470455 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
STAM1_HUMAN
|
||||||
| θ value | 7.84624e-12 (rank : 13) | NC score | 0.470409 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| θ value | 1.02475e-11 (rank : 14) | NC score | 0.463934 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 1.25267e-09 (rank : 15) | NC score | 0.429263 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
HGS_HUMAN
|
||||||
| θ value | 2.13673e-09 (rank : 16) | NC score | 0.431271 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.027153 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
TBCD1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.042559 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
CABYR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.070918 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.014089 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.013304 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
SRTD2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.043815 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.030663 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
PDCL3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.036516 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
KCC1G_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.002663 (rank : 49) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NX5, Q86UH5, Q9Y3J7 | Gene names | CAMK1G, VWS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type 1G (EC 2.7.11.17) (CaM kinase IG) (CaM kinase I gamma) (CaMKI gamma) (CaMKI-gamma) (CaM- KI gamma) (CaMKIG) (CaMK-like CREB kinase III) (CLICK III). | |||||
|
PP4R1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.019862 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
TITIN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.002864 (rank : 48) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
GORS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.028897 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.021918 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
VDP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.022347 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
MYH10_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.012271 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
MYH10_MOUSE
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.012482 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.012205 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
TMCO3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.018100 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UWJ1, Q5JSB1, Q6NUN1, Q8NG29, Q8TCI6, Q96EA6, Q9NWT2 | Gene names | TMCO3, C13orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor (Putative LAG1-interacting protein). | |||||
|
CND2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.014234 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C156, Q8BY01, Q8VDM9 | Gene names | Ncaph, Brrn, Brrn1, Caph | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (mCAP-H) (XCAP-H homolog). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.011561 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
I17RA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.012409 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60943 | Gene names | IL17ra, Il17r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor A precursor (IL-17 receptor). | |||||
|
MYPT1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.003694 (rank : 47) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
HCLS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.044068 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
HOME1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.009327 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
NCOA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.016026 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
NFIA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.012401 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12857, Q9H3X9, Q9P2A9 | Gene names | NFIA, KIAA1439 | |||
|
Domain Architecture |
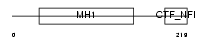
|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
NFIA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.011817 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02780, P70250, P70251, Q3UUZ2, Q61960, Q8VBT5, Q9R1G5 | Gene names | Nfia | |||
|
Domain Architecture |
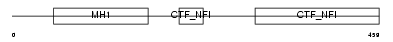
|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
AP1G1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.097838 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.094341 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.084586 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.089970 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
STAC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.051057 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MF2, Q96HU5 | Gene names | STAC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STAC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.051833 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
TM1L1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q923U0, Q99KE0, Q9D6Y5 | Gene names | Tom1l1, Srcasm | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.992571 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.910587 (rank : 3) | θ value | 2.03627e-60 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
TOM1_HUMAN
|
||||||
| NC score | 0.909091 (rank : 4) | θ value | 5.0155e-59 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60784, Q5TIJ6 | Gene names | TOM1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.675196 (rank : 5) | θ value | 2.35696e-16 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.656398 (rank : 6) | θ value | 3.40345e-15 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GGA1_MOUSE
|
||||||
| NC score | 0.654495 (rank : 7) | θ value | 6.85773e-16 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R0H9, Q3U2N1 | Gene names | Gga1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA1_HUMAN
|
||||||
| NC score | 0.652311 (rank : 8) | θ value | 1.16975e-15 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UJY5, Q5TG07, Q9BW94, Q9UG00, Q9UGW0, Q9UGW1 | Gene names | GGA1 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA1 (Golgi-localized, gamma ear-containing, ARF-binding protein 1) (Gamma-adaptin-related protein 1). | |||||
|
GGA2_HUMAN
|
||||||
| NC score | 0.651067 (rank : 9) | θ value | 6.20254e-17 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9UJY4, O14564, Q9NYN2, Q9UPS2 | Gene names | GGA2, KIAA1080 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2) (VHS domain and ear domain of gamma-adaptin) (Vear). | |||||
|
GGA2_MOUSE
|
||||||
| NC score | 0.645011 (rank : 10) | θ value | 3.40345e-15 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P5E6, Q3U374, Q6PFX3, Q6ZPY8, Q8BM76, Q9DC15 | Gene names | Gga2, Kiaa1080 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA2 (Golgi-localized, gamma ear-containing, ARF-binding protein 2) (Gamma-adaptin-related protein 2). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.485835 (rank : 11) | θ value | 4.91457e-14 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
STAM2_MOUSE
|
||||||
| NC score | 0.470455 (rank : 12) | θ value | 1.2105e-12 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 285 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O88811, Q8C8Y4 | Gene names | Stam2, Hbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2) (Hrs-binding protein). | |||||
|
STAM1_HUMAN
|
||||||
| NC score | 0.470409 (rank : 13) | θ value | 7.84624e-12 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q92783, Q8N6D9 | Gene names | STAM, STAM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
STAM1_MOUSE
|
||||||
| NC score | 0.463934 (rank : 14) | θ value | 1.02475e-11 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P70297 | Gene names | Stam, Stam1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 1 (STAM-1). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.431271 (rank : 15) | θ value | 2.13673e-09 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.429263 (rank : 16) | θ value | 1.25267e-09 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
AP1G1_HUMAN
|
||||||
| NC score | 0.097838 (rank : 17) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43747, O75709, O75842, Q9UG09, Q9Y3U4 | Gene names | AP1G1, ADTG, CLAPG1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G1_MOUSE
|
||||||
| NC score | 0.094341 (rank : 18) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22892 | Gene names | Ap1g1, Adtg, Clapg1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-1 (Adapter-related protein complex 1 gamma- 1 subunit) (Gamma-adaptin) (Adaptor protein complex AP-1 gamma-1 subunit) (Golgi adaptor HA1/AP1 adaptin subunit gamma-1) (Clathrin assembly protein complex 1 gamma-1 large chain). | |||||
|
AP1G2_MOUSE
|
||||||
| NC score | 0.089970 (rank : 19) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88512 | Gene names | Ap1g2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
AP1G2_HUMAN
|
||||||
| NC score | 0.084586 (rank : 20) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75843, O75504 | Gene names | AP1G2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-1 complex subunit gamma-2 (Adapter-related protein complex 1 gamma- 2 subunit) (Gamma2-adaptin) (Adaptor protein complex AP-1 gamma-2 subunit) (G2ad). | |||||
|
CABYR_HUMAN
|
||||||
| NC score | 0.070918 (rank : 21) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75952, Q8WXW5, Q9HAY3, Q9HAY4, Q9HAY5, Q9HCY9 | Gene names | CABYR, CBP86, FSP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-binding tyrosine phosphorylation-regulated protein (Calcium- binding protein 86) (Testis-specific calcium-binding protein CBP86) (Fibrousheathin-2) (FSP-2). | |||||
|
STAC3_MOUSE
|
||||||
| NC score | 0.051833 (rank : 22) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BZ71 | Gene names | Stac3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
STAC3_HUMAN
|
||||||
| NC score | 0.051057 (rank : 23) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96MF2, Q96HU5 | Gene names | STAC3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and cysteine-rich domain-containing protein 3. | |||||
|
HCLS1_HUMAN
|
||||||
| NC score | 0.044068 (rank : 24) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14317 | Gene names | HCLS1, HS1 | |||
|
Domain Architecture |

|
|||||
| Description | Hematopoietic lineage cell-specific protein (Hematopoietic cell- specific LYN substrate 1) (LckBP1) (p75). | |||||
|
SRTD2_HUMAN
|
||||||
| NC score | 0.043815 (rank : 25) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 62 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14140 | Gene names | SERTAD2, KIAA0127 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SERTA domain-containing protein 2 (Transcriptional regulator interacting with the PHD-bromodomain 2) (TRIP-Br2). | |||||
|
TBCD1_MOUSE
|
||||||
| NC score | 0.042559 (rank : 26) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q60949, Q80TJ9, Q923F8 | Gene names | Tbc1d1, Kiaa1108, Tbc1 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 1. | |||||
|
PDCL3_MOUSE
|
||||||
| NC score | 0.036516 (rank : 27) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVF2, Q3TH06, Q99JX2, Q9D0W3 | Gene names | Pdcl3, Viaf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosducin-like protein 3 (Viral IAP-associated factor 1) (VIAF-1). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.030663 (rank : 28) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
GORS2_HUMAN
|
||||||
| NC score | 0.028897 (rank : 29) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H8Y8, Q96I74, Q96K84, Q9H946, Q9UFW4 | Gene names | GORASP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55) (p59) (Golgi phosphoprotein 6) (GOLPH6). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.027153 (rank : 30) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
VDP_HUMAN
|
||||||
| NC score | 0.022347 (rank : 31) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60763 | Gene names | VDP | |||
|
Domain Architecture |

|
|||||
| Description | General vesicular transport factor p115 (Transcytosis-associated protein) (TAP) (Vesicle docking protein). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.021918 (rank : 32) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
PP4R1_HUMAN
|
||||||
| NC score | 0.019862 (rank : 33) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8TF05, Q99774, Q9UNQ7 | Gene names | PPP4R1, MEG1, PP4R1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein phosphatase 4 regulatory subunit 1. | |||||
|
TMCO3_HUMAN
|
||||||
| NC score | 0.018100 (rank : 34) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6UWJ1, Q5JSB1, Q6NUN1, Q8NG29, Q8TCI6, Q96EA6, Q9NWT2 | Gene names | TMCO3, C13orf11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane and coiled-coil domain-containing protein 3 precursor (Putative LAG1-interacting protein). | |||||
|
NCOA6_MOUSE
|
||||||
| NC score | 0.016026 (rank : 35) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1076 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9JL19, Q9JLT9 | Gene names | Ncoa6, Aib3, Prip, Rap250, Trbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 6 (Amplified in breast cancer protein 3) (Cancer-amplified transcriptional coactivator ASC-2) (Activating signal cointegrator 2) (ASC-2) (Peroxisome proliferator-activated receptor-interacting protein) (PPAR-interacting protein) (Nuclear receptor-activating protein, 250 kDa) (Nuclear receptor coactivator RAP250) (NRC) (Thyroid hormone receptor-binding protein). | |||||
|
CND2_MOUSE
|
||||||
| NC score | 0.014234 (rank : 36) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8C156, Q8BY01, Q8VDM9 | Gene names | Ncaph, Brrn, Brrn1, Caph | |||
|
Domain Architecture |

|
|||||
| Description | Condensin complex subunit 2 (Non-SMC condensin I complex subunit H) (Barren homolog protein 1) (Chromosome-associated protein H) (mCAP-H) (XCAP-H homolog). | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.014089 (rank : 37) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.013304 (rank : 38) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
MYH10_MOUSE
|
||||||
| NC score | 0.012482 (rank : 39) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1612 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q61879, Q5SV63 | Gene names | Myh10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
I17RA_MOUSE
|
||||||
| NC score | 0.012409 (rank : 40) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60943 | Gene names | IL17ra, Il17r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-17 receptor A precursor (IL-17 receptor). | |||||
|
NFIA_HUMAN
|
||||||
| NC score | 0.012401 (rank : 41) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12857, Q9H3X9, Q9P2A9 | Gene names | NFIA, KIAA1439 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
MYH10_HUMAN
|
||||||
| NC score | 0.012271 (rank : 42) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P35580, Q16087 | Gene names | MYH10 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.012205 (rank : 43) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
NFIA_MOUSE
|
||||||
| NC score | 0.011817 (rank : 44) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02780, P70250, P70251, Q3UUZ2, Q61960, Q8VBT5, Q9R1G5 | Gene names | Nfia | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 A-type (Nuclear factor 1/A) (NF1-A) (NFI-A) (NF-I/A) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.011561 (rank : 45) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
HOME1_MOUSE
|
||||||
| NC score | 0.009327 (rank : 46) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 630 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z2Y3, Q8K3E1, Q8K4M8, Q9Z0E9, Q9Z216 | Gene names | Homer1, Vesl1 | |||
|
Domain Architecture |

|
|||||
| Description | Homer protein homolog 1 (VASP/Ena-related gene up-regulated during seizure and LTP) (Vesl-1). | |||||
|
MYPT1_HUMAN
|
||||||
| NC score | 0.003694 (rank : 47) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O14974, Q86WU3, Q8NFR6, Q9BYH0 | Gene names | PPP1R12A, MBS, MYPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 12A (Myosin phosphatase- targeting subunit 1) (Myosin phosphatase target subunit 1) (Protein phosphatase myosin-binding subunit). | |||||
|
TITIN_HUMAN
|
||||||
| NC score | 0.002864 (rank : 48) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 2923 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8WZ42, Q10465, Q10466, Q15598, Q2XUS3, Q32Q60, Q4U1Z6, Q6NSG0, Q6PDB1, Q6PJP0, Q7KYM2, Q7KYN4, Q7KYN5, Q7LDM3, Q7Z2X3, Q8TCG8, Q8WZ51, Q8WZ52, Q8WZ53, Q8WZB3, Q92761, Q92762, Q9UD97, Q9UP84, Q9Y6L9 | Gene names | TTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Titin (EC 2.7.11.1) (Connectin) (Rhabdomyosarcoma antigen MU-RMS- 40.14). | |||||
|
KCC1G_HUMAN
|
||||||
| NC score | 0.002663 (rank : 49) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96NX5, Q86UH5, Q9Y3J7 | Gene names | CAMK1G, VWS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium/calmodulin-dependent protein kinase type 1G (EC 2.7.11.17) (CaM kinase IG) (CaM kinase I gamma) (CaMKI gamma) (CaMKI-gamma) (CaM- KI gamma) (CaMKIG) (CaMK-like CREB kinase III) (CLICK III). | |||||