Please be patient as the page loads
|
STRC_HUMAN
|
||||||
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
STRC_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
STRC_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.981568 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
OTOAN_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 3) | NC score | 0.518795 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTW8, Q8NA86, Q96M76 | Gene names | OTOA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
OTOAN_MOUSE
|
||||||
| θ value | 1.02238e-19 (rank : 4) | NC score | 0.505861 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
NRCAM_HUMAN
|
||||||
| θ value | 0.163984 (rank : 5) | NC score | 0.014841 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92823, O15051, O15179, Q9UHI3, Q9UHI4 | Gene names | NRCAM, KIAA0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (hBravo). | |||||
|
FOXH1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.030617 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
PR285_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.040794 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
VCAM1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.021569 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
STAT6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.030632 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
ATS8_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.011264 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UP79, Q9NZS0 | Gene names | ADAMTS8, METH2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 8) (ADAM-TS 8) (ADAM-TS8) (METH-2) (METH- 8). | |||||
|
ITPR1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.027617 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
ITPR1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.027627 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.029319 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
LPLC4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.038163 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59827, Q5TDX6 | Gene names | LPLUNC4, C20orf186 | |||
|
Domain Architecture |
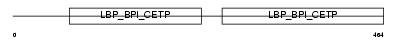
|
|||||
| Description | Long palate, lung and nasal epithelium carcinoma-associated protein 4 (Ligand-binding protein RY2G5). | |||||
|
CN159_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.025864 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3D6, Q86SW3, Q86SX8, Q86SX9, Q86T08, Q86TV5, Q96GW5, Q9H7G0, Q9H8Y9, Q9H9W6 | Gene names | C14orf159 | |||
|
Domain Architecture |
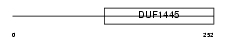
|
|||||
| Description | UPF0317 protein C14orf159, mitochondrial precursor. | |||||
|
PHLPL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.008995 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
TOM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.017577 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
ATF5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.024104 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
CP1A1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.009471 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00184, Q61455 | Gene names | Cyp1a1, Cyp1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1). | |||||
|
PCDGD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.007705 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
POLS2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.004659 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
CCNG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.020388 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |
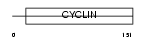
|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.020370 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |
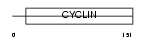
|
|||||
| Description | Cyclin-G2. | |||||
|
IGHD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.013218 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01880 | Gene names | IGHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region. | |||||
|
SF04_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.016940 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
AMOT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.007558 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
ATG4D_HUMAN
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.011581 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86TL0 | Gene names | ATG4D, APG4D, AUTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine protease ATG4D (EC 3.4.22.-) (Autophagy-related protein 4 homolog D) (Autophagin-4) (Autophagy-related cysteine endopeptidase 4) (AUT-like 4 cysteine endopeptidase). | |||||
|
MYNN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.000039 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MD8, Q6P1G7, Q8BT55, Q922I4, Q9CSA0, Q9CXJ8 | Gene names | Mynn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myoneurin. | |||||
|
SSPO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.005670 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
Z3H7B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.015138 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |
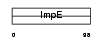
|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
WDR23_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.006685 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VU6, Q3U836, Q8JZV8 | Gene names | Wdr23, D14Ucla1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 23. | |||||
|
ZN236_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.000192 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
STRC_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q7RTU9 | Gene names | STRC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
STRC_MOUSE
|
||||||
| NC score | 0.981568 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VIM6 | Gene names | Strc | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stereocilin precursor. | |||||
|
OTOAN_HUMAN
|
||||||
| NC score | 0.518795 (rank : 3) | θ value | 2.06002e-20 (rank : 3) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTW8, Q8NA86, Q96M76 | Gene names | OTOA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
OTOAN_MOUSE
|
||||||
| NC score | 0.505861 (rank : 4) | θ value | 1.02238e-19 (rank : 4) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K561 | Gene names | Otoa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Otoancorin precursor. | |||||
|
PR285_HUMAN
|
||||||
| NC score | 0.040794 (rank : 5) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYK8, Q3C2G2, Q4VXQ1, Q8TEF3, Q96ND3, Q9C094 | Gene names | PRIC285, KIAA1769 | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal proliferator-activated receptor A-interacting complex 285 kDa protein (EC 3.6.1.-) (ATP-dependent helicase PRIC285) (PPAR-alpha- interacting complex protein 285) (PPAR-gamma DBD-interacting protein 1) (PDIP1). | |||||
|
LPLC4_HUMAN
|
||||||
| NC score | 0.038163 (rank : 6) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59827, Q5TDX6 | Gene names | LPLUNC4, C20orf186 | |||
|
Domain Architecture |

|
|||||
| Description | Long palate, lung and nasal epithelium carcinoma-associated protein 4 (Ligand-binding protein RY2G5). | |||||
|
STAT6_HUMAN
|
||||||
| NC score | 0.030632 (rank : 7) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42226 | Gene names | STAT6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and activator of transcription 6 (IL-4 Stat). | |||||
|
FOXH1_HUMAN
|
||||||
| NC score | 0.030617 (rank : 8) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O75593 | Gene names | FOXH1, FAST1, FAST2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein H1 (Forkhead activin signal transducer 1) (Fast- 1) (hFAST-1) (Forkhead activin signal transducer 2) (Fast-2). | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.029319 (rank : 9) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
ITPR1_MOUSE
|
||||||
| NC score | 0.027627 (rank : 10) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11881, P20943, Q99LG5 | Gene names | Itpr1, Insp3r, Pcd6, Pcp1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (Inositol 1,4,5-trisphosphate-binding protein P400) (Purkinje cell protein 1) (Protein PCD-6). | |||||
|
ITPR1_HUMAN
|
||||||
| NC score | 0.027617 (rank : 11) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14643, Q14660, Q99897 | Gene names | ITPR1, INSP3R1 | |||
|
Domain Architecture |

|
|||||
| Description | Inositol 1,4,5-trisphosphate receptor type 1 (Type 1 inositol 1,4,5- trisphosphate receptor) (Type 1 InsP3 receptor) (IP3 receptor isoform 1) (InsP3R1) (IP3R). | |||||
|
CN159_HUMAN
|
||||||
| NC score | 0.025864 (rank : 12) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3D6, Q86SW3, Q86SX8, Q86SX9, Q86T08, Q86TV5, Q96GW5, Q9H7G0, Q9H8Y9, Q9H9W6 | Gene names | C14orf159 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0317 protein C14orf159, mitochondrial precursor. | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.024104 (rank : 13) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
VCAM1_MOUSE
|
||||||
| NC score | 0.021569 (rank : 14) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P29533 | Gene names | Vcam1, Vcam-1 | |||
|
Domain Architecture |

|
|||||
| Description | Vascular cell adhesion protein 1 precursor (V-CAM 1). | |||||
|
CCNG2_HUMAN
|
||||||
| NC score | 0.020388 (rank : 15) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16589 | Gene names | CCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
CCNG2_MOUSE
|
||||||
| NC score | 0.020370 (rank : 16) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08918, O35612 | Gene names | Ccng2 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-G2. | |||||
|
TOM1_MOUSE
|
||||||
| NC score | 0.017577 (rank : 17) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O88746, Q3V4C6, Q9D120 | Gene names | Tom1 | |||
|
Domain Architecture |

|
|||||
| Description | Target of Myb protein 1. | |||||
|
SF04_HUMAN
|
||||||
| NC score | 0.016940 (rank : 18) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWZ8, O60378, Q6P3X9, Q8TCQ4, Q8WWT4, Q8WWT5, Q9NTG3 | Gene names | SF4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor 4 (RNA-binding protein RBP). | |||||
|
Z3H7B_HUMAN
|
||||||
| NC score | 0.015138 (rank : 19) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UGR2, Q5TFX9, Q8TBT9, Q9H8B6, Q9UGQ9, Q9UGR0, Q9UGR1, Q9UK03, Q9UPW9 | Gene names | ZC3H7B, KIAA1031 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH-type domain-containing protein 7B (Rotavirus 'X'- associated non-structural protein) (RoXaN). | |||||
|
NRCAM_HUMAN
|
||||||
| NC score | 0.014841 (rank : 20) | θ value | 0.163984 (rank : 5) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92823, O15051, O15179, Q9UHI3, Q9UHI4 | Gene names | NRCAM, KIAA0343 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal cell adhesion molecule precursor (Nr-CAM) (NgCAM-related cell adhesion molecule) (Ng-CAM-related) (hBravo). | |||||
|
IGHD_HUMAN
|
||||||
| NC score | 0.013218 (rank : 21) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01880 | Gene names | IGHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region. | |||||
|
ATG4D_HUMAN
|
||||||
| NC score | 0.011581 (rank : 22) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86TL0 | Gene names | ATG4D, APG4D, AUTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine protease ATG4D (EC 3.4.22.-) (Autophagy-related protein 4 homolog D) (Autophagin-4) (Autophagy-related cysteine endopeptidase 4) (AUT-like 4 cysteine endopeptidase). | |||||
|
ATS8_HUMAN
|
||||||
| NC score | 0.011264 (rank : 23) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UP79, Q9NZS0 | Gene names | ADAMTS8, METH2 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-8 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 8) (ADAM-TS 8) (ADAM-TS8) (METH-2) (METH- 8). | |||||
|
CP1A1_MOUSE
|
||||||
| NC score | 0.009471 (rank : 24) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P00184, Q61455 | Gene names | Cyp1a1, Cyp1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 1A1 (EC 1.14.14.1) (CYPIA1) (P450-P1). | |||||
|
PHLPL_HUMAN
|
||||||
| NC score | 0.008995 (rank : 25) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZVD8, Q9NV17, Q9Y2E3 | Gene names | PHLPPL, KIAA0931 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat protein phosphatase-like (EC 3.1.3.16). | |||||
|
PCDGD_HUMAN
|
||||||
| NC score | 0.007705 (rank : 26) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
AMOT_HUMAN
|
||||||
| NC score | 0.007558 (rank : 27) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 712 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q4VCS5, Q504X5, Q9HD27, Q9UPT1 | Gene names | AMOT, KIAA1071 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Angiomotin. | |||||
|
WDR23_MOUSE
|
||||||
| NC score | 0.006685 (rank : 28) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91VU6, Q3U836, Q8JZV8 | Gene names | Wdr23, D14Ucla1 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 23. | |||||
|
SSPO_MOUSE
|
||||||
| NC score | 0.005670 (rank : 29) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG65 | Gene names | Sspo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SCO-spondin precursor. | |||||
|
POLS2_HUMAN
|
||||||
| NC score | 0.004659 (rank : 30) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
ZN236_HUMAN
|
||||||
| NC score | 0.000192 (rank : 31) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UL36, Q9UL37 | Gene names | ZNF236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 236. | |||||
|
MYNN_MOUSE
|
||||||
| NC score | 0.000039 (rank : 32) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 32 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99MD8, Q6P1G7, Q8BT55, Q922I4, Q9CSA0, Q9CXJ8 | Gene names | Mynn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myoneurin. | |||||