Please be patient as the page loads
|
MCPH1_HUMAN
|
||||||
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MCPH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MCPH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.931552 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 1.80886e-08 (rank : 3) | NC score | 0.243378 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 1.43324e-05 (rank : 4) | NC score | 0.212084 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
TOPB1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 5) | NC score | 0.197331 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
BARD1_HUMAN
|
||||||
| θ value | 0.000786445 (rank : 6) | NC score | 0.110510 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BARD1_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 7) | NC score | 0.103884 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 8) | NC score | 0.171037 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
ZBTB5_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 9) | NC score | 0.029034 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 801 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQG0, Q3TVR7, Q6GV09, Q8BIB5 | Gene names | Zbtb5, Kiaa0354 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 5 (Transcription factor ZNF-POZ). | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.074690 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 11) | NC score | 0.087591 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
ZBTB5_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 12) | NC score | 0.026210 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15062 | Gene names | ZBTB5, KIAA0354 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger and BTB domain-containing protein 5. | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 13) | NC score | 0.044545 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.042571 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
MTMR3_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.034800 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.043219 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.043125 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.041271 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 19) | NC score | 0.034547 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.050760 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.057782 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
SEPT6_MOUSE
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.029385 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R1T4, Q91XH2, Q9CZ94 | Gene names | Sept6 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-6. | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.005103 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CDKL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.004209 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92772 | Gene names | CDKL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE) (Protein kinase p56 KKIAMRE). | |||||
|
GGA3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.028074 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
PARG_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.040817 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
CIR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.069661 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
REPS2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.030950 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |
|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.031808 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.031897 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
REST_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.025316 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SEPT6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.023364 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-6. | |||||
|
BPTF_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.029686 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
CNGA1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.013531 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.027042 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PCDA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.045998 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDA2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.045653 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDA3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.045784 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDA4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.046535 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.045629 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDA6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.046322 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.046157 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDA8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.046447 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.046659 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDAA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.046488 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
PCDAB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.045864 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
PCDAC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.046226 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDAD_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.046026 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDC1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.041357 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
PCDC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.039975 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
ABTAP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.043305 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
ABTAP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.042518 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
CF182_MOUSE
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.031922 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
ENAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.025484 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
FA21C_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.034643 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
FA48A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.028792 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM7, Q71RF3, Q9Y6A6 | Gene names | FAM48A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
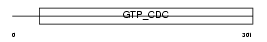
PYR5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.029887 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11172, O00758, O00759, O00760, Q16862, Q9H3Q2, Q9UG49 | Gene names | UMPS | |||
|
Domain Architecture |
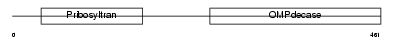
|
|||||
| Description | Uridine 5'-monophosphate synthase (UMP synthase) [Includes: Orotate phosphoribosyltransferase (EC 2.4.2.10) (OPRTase); Orotidine 5'- phosphate decarboxylase (EC 4.1.1.23) (OMPdecase)]. | |||||
|
TAU_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.021537 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
BRCA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.019085 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.024544 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DPOLZ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.016971 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
INVS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.012799 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
KIF1C_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.010331 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.018513 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
LIPA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.012553 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |
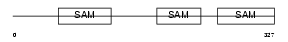
|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
PCDA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.044708 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
SNUT1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.022482 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
MMTA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.022907 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
MRIP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.030183 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PTN13_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.003926 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.028035 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
SYTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.009716 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.021374 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TNR8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.015659 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
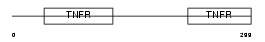
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
CACB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012401 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.011989 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |
|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
CASC5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.022208 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.029221 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
GLGB_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.014366 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D6Y9 | Gene names | Gbe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.019327 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
K0460_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.024823 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
K0460_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.023949 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
KIF1C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.008908 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.031024 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MRIP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.028871 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.050830 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
MCPH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q8NEM0, Q66GU1, Q9H9C7 | Gene names | MCPH1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
MCPH1_MOUSE
|
||||||
| NC score | 0.931552 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7TT79, Q8BNI9, Q8C9I7 | Gene names | Mcph1 | |||
|
Domain Architecture |

|
|||||
| Description | Microcephalin. | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.243378 (rank : 3) | θ value | 1.80886e-08 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.212084 (rank : 4) | θ value | 1.43324e-05 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
TOPB1_HUMAN
|
||||||
| NC score | 0.197331 (rank : 5) | θ value | 2.44474e-05 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q92547, Q7LGC1, Q9UEB9 | Gene names | TOPBP1, KIAA0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase 2-binding protein 1 (DNA topoisomerase II-binding protein 1) (DNA topoisomerase IIbeta-binding protein 1) (TopBP1). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.171037 (rank : 6) | θ value | 0.00665767 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
BARD1_HUMAN
|
||||||
| NC score | 0.110510 (rank : 7) | θ value | 0.000786445 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99728, O43574 | Gene names | BARD1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
BARD1_MOUSE
|
||||||
| NC score | 0.103884 (rank : 8) | θ value | 0.00175202 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 411 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O70445 | Gene names | Bard1 | |||
|
Domain Architecture |

|
|||||
| Description | BRCA1-associated RING domain protein 1 (BARD-1). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.087591 (rank : 9) | θ value | 0.0563607 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.074690 (rank : 10) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.069661 (rank : 11) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.057782 (rank : 12) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.050830 (rank : 13) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 1 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.050760 (rank : 14) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.046659 (rank : 15) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDA4_HUMAN
|
||||||
| NC score | 0.046535 (rank : 16) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDAA_HUMAN
|
||||||
| NC score | 0.046488 (rank : 17) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
PCDA8_HUMAN
|
||||||
| NC score | 0.046447 (rank : 18) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDA6_HUMAN
|
||||||
| NC score | 0.046322 (rank : 19) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDAC_HUMAN
|
||||||
| NC score | 0.046226 (rank : 20) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.046157 (rank : 21) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDAD_HUMAN
|
||||||
| NC score | 0.046026 (rank : 22) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDA1_HUMAN
|
||||||
| NC score | 0.045998 (rank : 23) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDAB_HUMAN
|
||||||
| NC score | 0.045864 (rank : 24) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
PCDA3_HUMAN
|
||||||
| NC score | 0.045784 (rank : 25) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDA2_HUMAN
|
||||||
| NC score | 0.045653 (rank : 26) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.045629 (rank : 27) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDA4_MOUSE
|
||||||
| NC score | 0.044708 (rank : 28) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.044545 (rank : 29) | θ value | 0.0961366 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
ABTAP_HUMAN
|
||||||
| NC score | 0.043305 (rank : 30) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H501, Q86X92, Q9H9Q5, Q9HA35, Q9NX93, Q9P1S6 | Gene names | ABTAP, C20orf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.043219 (rank : 31) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.043125 (rank : 32) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.042571 (rank : 33) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
ABTAP_MOUSE
|
||||||
| NC score | 0.042518 (rank : 34) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3V1V3 | Gene names | Abtap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ABT1-associated protein. | |||||
|
PCDC1_HUMAN
|
||||||
| NC score | 0.041357 (rank : 35) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.041271 (rank : 36) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.040817 (rank : 37) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PCDC2_HUMAN
|
||||||
| NC score | 0.039975 (rank : 38) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
MTMR3_HUMAN
|
||||||
| NC score | 0.034800 (rank : 39) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13615, Q9NYN5, Q9NYN6, Q9UDX6, Q9UEG3 | Gene names | MTMR3, KIAA0371, ZFYVE10 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 3 (EC 3.1.3.48) (FYVE domain-containing dual specificity protein phosphatase 1) (FYVE-DSP1) (Zinc finger FYVE domain-containing protein 10). | |||||
|
FA21C_HUMAN
|
||||||
| NC score | 0.034643 (rank : 40) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y4E1, Q5SQU4, Q5SQU5, Q7L521, Q9UG79 | Gene names | FAM21C, KIAA0592 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM21C. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.034547 (rank : 41) | θ value | 0.365318 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
CF182_MOUSE
|
||||||
| NC score | 0.031922 (rank : 42) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 551 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8VDS7, Q9CZE0, Q9D5A1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf182 homolog. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.031897 (rank : 43) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.031808 (rank : 44) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.031024 (rank : 45) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
REPS2_MOUSE
|
||||||
| NC score | 0.030950 (rank : 46) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80XA6 | Gene names | Reps2, Pob1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
MRIP_HUMAN
|
||||||
| NC score | 0.030183 (rank : 47) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 578 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6WCQ1, Q3KQZ5, Q5FB94, Q6DHW2, Q7Z5Y2, Q8N390, Q96G40 | Gene names | MRIP, KIAA0864, RHOIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (M-RIP) (RIP3) (p116Rip). | |||||
|
PYR5_HUMAN
|
||||||
| NC score | 0.029887 (rank : 48) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11172, O00758, O00759, O00760, Q16862, Q9H3Q2, Q9UG49 | Gene names | UMPS | |||
|
Domain Architecture |

|
|||||
| Description | Uridine 5'-monophosphate synthase (UMP synthase) [Includes: Orotate phosphoribosyltransferase (EC 2.4.2.10) (OPRTase); Orotidine 5'- phosphate decarboxylase (EC 4.1.1.23) (OMPdecase)]. | |||||
|
BPTF_HUMAN
|
||||||
| NC score | 0.029686 (rank : 49) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12830, Q6NX67, Q7Z7D6, Q9UIG2 | Gene names | FALZ, BPTF, FAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleosome remodeling factor subunit BPTF (Bromodomain and PHD finger- containing transcription factor) (Fetal Alzheimer antigen) (Fetal Alz- 50 clone 1 protein). | |||||
|
SEPT6_MOUSE
|
||||||
| NC score | 0.029385 (rank : 50) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9R1T4, Q91XH2, Q9CZ94 | Gene names | Sept6 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.029221 (rank : 51) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
ZBTB5_MOUSE
|
||||||
| NC score | 0.029034 (rank : 52) | θ value | 0.00665767 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 801 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQG0, Q3TVR7, Q6GV09, Q8BIB5 | Gene names | Zbtb5, Kiaa0354 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 5 (Transcription factor ZNF-POZ). | |||||
|
MRIP_MOUSE
|
||||||
| NC score | 0.028871 (rank : 53) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P97434, Q3TBR5, Q3TC85, Q3TRS0, Q3U1Y8, Q3U3E4, Q5SWZ3, Q5SWZ4, Q5SWZ7, Q6P559, Q80YC8 | Gene names | Mrip, Kiaa0864, Rhoip3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin phosphatase Rho-interacting protein (Rho-interacting protein 3) (p116Rip) (RIP3). | |||||
|
FA48A_HUMAN
|
||||||
| NC score | 0.028792 (rank : 54) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NEM7, Q71RF3, Q9Y6A6 | Gene names | FAM48A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM48A (p38-interacting protein) (p38IP). | |||||
|
GGA3_MOUSE
|
||||||
| NC score | 0.028074 (rank : 55) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BMI3, Q80U71 | Gene names | Gga3, Kiaa0154 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.028035 (rank : 56) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.027042 (rank : 57) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
ZBTB5_HUMAN
|
||||||
| NC score | 0.026210 (rank : 58) | θ value | 0.0563607 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 811 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15062 | Gene names | ZBTB5, KIAA0354 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 5. | |||||
|
ENAM_MOUSE
|
||||||
| NC score | 0.025484 (rank : 59) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55196 | Gene names | Enam | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Enamelin precursor. | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.025316 (rank : 60) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
K0460_HUMAN
|
||||||
| NC score | 0.024823 (rank : 61) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VT52, O75048, Q5VT53, Q6MZL4, Q86XD2 | Gene names | KIAA0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.024544 (rank : 62) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.023949 (rank : 63) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
SEPT6_HUMAN
|
||||||
| NC score | 0.023364 (rank : 64) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
MMTA2_HUMAN
|
||||||
| NC score | 0.022907 (rank : 65) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BU76, Q6P5Y0, Q6ZTZ6, Q6ZWA6, Q8IZH3 | Gene names | MMTAG2, C1orf35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple myeloma tumor-associated protein 2 (hMMTAG2). | |||||
|
SNUT1_MOUSE
|
||||||
| NC score | 0.022482 (rank : 66) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z315, Q8K155, Q9R1I9, Q9R270 | Gene names | Sart1, Haf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U4/U6.U5 tri-snRNP-associated protein 1 (Squamous cell carcinoma antigen recognized by T-cells 1) (SART-1) (mSART-1) (Hypoxia- associated factor). | |||||
|
CASC5_HUMAN
|
||||||
| NC score | 0.022208 (rank : 67) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 376 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NG31, Q8NHE1, Q8WXA6, Q9HCK2, Q9NR92 | Gene names | CASC5, KIAA1570 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASC5 (Cancer susceptibility candidate gene 5 protein) (ALL1- fused gene from chromosome 15q14) (AF15q14) (D40/AF15q14 protein). | |||||
|
TAU_HUMAN
|
||||||
| NC score | 0.021537 (rank : 68) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 448 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10636, P18518, Q14799, Q15549, Q15550, Q15551, Q9UDJ3, Q9UMH0, Q9UQ96 | Gene names | MAPT, MTBT1, TAU | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.021374 (rank : 69) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.019327 (rank : 70) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
BRCA2_MOUSE
|
||||||
| NC score | 0.019085 (rank : 71) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97929, O35922, P97383 | Gene names | Brca2, Fancd1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein homolog (Fanconi anemia group D1 protein homolog). | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.018513 (rank : 72) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
DPOLZ_MOUSE
|
||||||
| NC score | 0.016971 (rank : 73) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61493, Q9JMD6, Q9QWX6 | Gene names | Rev3l, Polz, Sez4 | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase zeta catalytic subunit (EC 2.7.7.7) (Seizure-related protein 4). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.015659 (rank : 74) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
GLGB_MOUSE
|
||||||
| NC score | 0.014366 (rank : 75) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D6Y9 | Gene names | Gbe1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1,4-alpha-glucan branching enzyme (EC 2.4.1.18) (Glycogen branching enzyme) (Brancher enzyme). | |||||
|
CNGA1_MOUSE
|
||||||
| NC score | 0.013531 (rank : 76) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29974, Q60776 | Gene names | Cnga1, Cncg, Cncg1 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-gated cation channel alpha 1 (CNG channel alpha 1) (CNG-1) (CNG1) (Cyclic nucleotide-gated channel alpha 1) (Cyclic nucleotide-gated channel, photoreceptor) (Cyclic nucleotide-gated cation channel 1) (Rod photoreceptor cGMP-gated channel subunit alpha). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.012799 (rank : 77) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
LIPA4_HUMAN
|
||||||
| NC score | 0.012553 (rank : 78) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75335, O94971 | Gene names | PPFIA4, KIAA0897 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-4 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-4) (PTPRF-interacting protein alpha-4). | |||||
|
CACB3_HUMAN
|
||||||
| NC score | 0.012401 (rank : 79) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54284, Q13913 | Gene names | CACNB3, CACNLB3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3). | |||||
|
CACB3_MOUSE
|
||||||
| NC score | 0.011989 (rank : 80) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P54285 | Gene names | Cacnb3, Cacnlb3 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit beta-3 (CAB3) (Calcium channel voltage-dependent subunit beta 3) (CCHB3). | |||||
|
KIF1C_HUMAN
|
||||||
| NC score | 0.010331 (rank : 81) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43896, O75186 | Gene names | KIF1C, KIAA0706 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
SYTL2_HUMAN
|
||||||
| NC score | 0.009716 (rank : 82) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HCH5, Q2YDA7, Q8ND34, Q96BJ2, Q9H768, Q9NXM1 | Gene names | SYTL2, KIAA1597, SLP2 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptotagmin-like protein 2 (Exophilin-4). | |||||
|
KIF1C_MOUSE
|
||||||
| NC score | 0.008908 (rank : 83) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35071, Q5SX62 | Gene names | Kif1c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin-like protein KIF1C. | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.005103 (rank : 84) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CDKL2_HUMAN
|
||||||
| NC score | 0.004209 (rank : 85) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 838 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92772 | Gene names | CDKL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-dependent kinase-like 2 (EC 2.7.11.22) (Serine/threonine- protein kinase KKIAMRE) (Protein kinase p56 KKIAMRE). | |||||
|
PTN13_MOUSE
|
||||||
| NC score | 0.003926 (rank : 86) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64512, Q61494, Q62135, Q64499 | Gene names | Ptpn13, Ptp14 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 13 (EC 3.1.3.48) (Protein tyrosine phosphatase PTP-BL) (Protein-tyrosine phosphatase RIP) (protein tyrosine phosphatase DPZPTP) (PTP36). | |||||