Please be patient as the page loads
|
EYA2_HUMAN
|
||||||
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
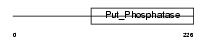
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EYA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.996046 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA1_HUMAN
|
||||||
| θ value | 1.40939e-178 (rank : 3) | NC score | 0.974414 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 3.5924e-174 (rank : 4) | NC score | 0.971986 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 3.47952e-169 (rank : 5) | NC score | 0.970280 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 2.33935e-157 (rank : 6) | NC score | 0.979375 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| θ value | 1.43579e-114 (rank : 7) | NC score | 0.978536 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| θ value | 2.37213e-109 (rank : 8) | NC score | 0.976038 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
TBX15_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.027179 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
TBX15_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.024483 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |
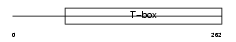
|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
K0141_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.025348 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCV6, Q6A0B8 | Gene names | Kiaa0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
CAPS2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.017923 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
STAM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.009440 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.002085 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
DAB2P_HUMAN
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.013287 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
HXB8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.011779 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17481, Q9H1I2 | Gene names | HOXB8, HOX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2D) (Hox-2.4). | |||||
|
HXB8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.011779 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09632 | Gene names | Hoxb8, Hox-2.4, Hoxb-8 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2.4). | |||||
|
GCM2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.012989 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75603 | Gene names | GCM2, GCMB | |||
|
Domain Architecture |
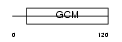
|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (hGCMb). | |||||
|
CAPS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.015617 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
DAB2P_MOUSE
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.012217 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
MSH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.012181 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.008330 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
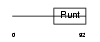
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 23) | NC score | 0.039682 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
SOX30_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.005316 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
SSRP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.006769 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
ZN575_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.000433 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
DEND_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.015307 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
IFI3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.012346 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |
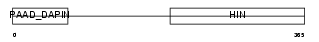
|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
EWS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.085977 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.085787 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
FUS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.084374 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
FUS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.095252 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| NC score | 0.996046 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.979375 (rank : 3) | θ value | 2.33935e-157 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| NC score | 0.978536 (rank : 4) | θ value | 1.43579e-114 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| NC score | 0.976038 (rank : 5) | θ value | 2.37213e-109 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.974414 (rank : 6) | θ value | 1.40939e-178 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.971986 (rank : 7) | θ value | 3.5924e-174 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.970280 (rank : 8) | θ value | 3.47952e-169 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.095252 (rank : 9) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.085977 (rank : 10) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.085787 (rank : 11) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.084374 (rank : 12) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.039682 (rank : 13) | θ value | 6.88961 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
TBX15_HUMAN
|
||||||
| NC score | 0.027179 (rank : 14) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96SF7, Q5T9S7 | Gene names | TBX15, TBX14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15). | |||||
|
K0141_MOUSE
|
||||||
| NC score | 0.025348 (rank : 15) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCV6, Q6A0B8 | Gene names | Kiaa0141 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0141. | |||||
|
TBX15_MOUSE
|
||||||
| NC score | 0.024483 (rank : 16) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O70306, O54840 | Gene names | Tbx15, Tbx14, Tbx8 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX15 (T-box protein 15) (MmTBx8). | |||||
|
CAPS2_HUMAN
|
||||||
| NC score | 0.017923 (rank : 17) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
CAPS2_MOUSE
|
||||||
| NC score | 0.015617 (rank : 18) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
DEND_MOUSE
|
||||||
| NC score | 0.015307 (rank : 19) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
DAB2P_HUMAN
|
||||||
| NC score | 0.013287 (rank : 20) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5VWQ8, Q8TDL2, Q96SE1, Q9C0C0 | Gene names | DAB2IP, AF9Q34, AIP1, KIAA1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein) (DAB2 interaction protein) (ASK-interacting protein 1). | |||||
|
GCM2_HUMAN
|
||||||
| NC score | 0.012989 (rank : 21) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75603 | Gene names | GCM2, GCMB | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (hGCMb). | |||||
|
IFI3_MOUSE
|
||||||
| NC score | 0.012346 (rank : 22) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
DAB2P_MOUSE
|
||||||
| NC score | 0.012217 (rank : 23) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q3UHC7, Q3TPD5, Q3UH44, Q6JTV1, Q80T97 | Gene names | Dab2ip, Kiaa1743 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disabled homolog 2-interacting protein (DAB2-interacting protein). | |||||
|
MSH3_MOUSE
|
||||||
| NC score | 0.012181 (rank : 24) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
HXB8_HUMAN
|
||||||
| NC score | 0.011779 (rank : 25) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17481, Q9H1I2 | Gene names | HOXB8, HOX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2D) (Hox-2.4). | |||||
|
HXB8_MOUSE
|
||||||
| NC score | 0.011779 (rank : 26) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09632 | Gene names | Hoxb8, Hox-2.4, Hoxb-8 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2.4). | |||||
|
STAM2_HUMAN
|
||||||
| NC score | 0.009440 (rank : 27) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75886, Q7LDQ0, Q9UF58 | Gene names | STAM2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Signal transducing adapter molecule 2 (STAM-2). | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.008330 (rank : 28) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
SSRP1_MOUSE
|
||||||
| NC score | 0.006769 (rank : 29) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08943, Q3U9Z2, Q3UJ75, Q4V9U4, Q8CGA6 | Gene names | Ssrp1 | |||
|
Domain Architecture |

|
|||||
| Description | FACT complex subunit SSRP1 (Facilitates chromatin transcription complex subunit SSRP1) (Structure-specific recognition protein 1) (Recombination signal sequence recognition protein 1) (T160). | |||||
|
SOX30_HUMAN
|
||||||
| NC score | 0.005316 (rank : 30) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O94993, O94995, Q8IYX6 | Gene names | SOX30 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-30. | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.002085 (rank : 31) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
ZN575_HUMAN
|
||||||
| NC score | 0.000433 (rank : 32) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||