Please be patient as the page loads
|
MSH3_MOUSE
|
||||||
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MSH3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.992779 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH6_HUMAN
|
||||||
| θ value | 1.65806e-78 (rank : 3) | NC score | 0.855877 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 3.45727e-76 (rank : 4) | NC score | 0.862072 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH4_MOUSE
|
||||||
| θ value | 7.98882e-65 (rank : 5) | NC score | 0.870466 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
MSH4_HUMAN
|
||||||
| θ value | 1.36269e-64 (rank : 6) | NC score | 0.871615 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH2_HUMAN
|
||||||
| θ value | 4.38363e-63 (rank : 7) | NC score | 0.872142 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 8) | NC score | 0.872697 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH5_MOUSE
|
||||||
| θ value | 2.11701e-41 (rank : 9) | NC score | 0.823997 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_HUMAN
|
||||||
| θ value | 6.15952e-41 (rank : 10) | NC score | 0.822937 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
CCD41_MOUSE
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.016124 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.022741 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ASGR1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.024882 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |
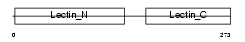
|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
GDE5_MOUSE
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.040474 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0L9, Q3TLV6, Q80TD5, Q8BKJ7, Q8BKW7, Q8CFW2, Q9D759 | Gene names | Gde5, Kiaa1434, Prei4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative glycerophosphodiester phosphodiesterase 5 (EC 3.1.-.-) (Preimplantation protein 4). | |||||
|
MYH14_MOUSE
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.012393 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.022158 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.015506 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
GDE5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.035050 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPB8, Q9BQL8, Q9NUX0 | Gene names | GDE5, KIAA1434 | |||
|
Domain Architecture |

|
|||||
| Description | Putative glycerophosphodiester phosphodiesterase 5 (EC 3.1.-.-). | |||||
|
NIBA_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.021228 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
MYH14_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.011023 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.011740 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
CP250_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.013459 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.012961 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.015726 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.015446 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SAMH1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.017075 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60710, Q91VK8 | Gene names | Samhd1, Mg11 | |||
|
Domain Architecture |
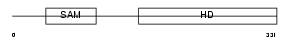
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Interferon-gamma- inducible protein Mg11). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.015725 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
EYA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.012181 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
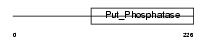
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.012118 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.011545 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.009136 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CT096_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.015682 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.013759 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
OLR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.008240 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78380, Q2PP00, Q7Z484 | Gene names | OLR1, LOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) (hLOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
TAC2N_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.010164 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |
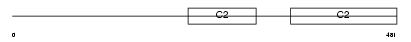
|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
DTNA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.009446 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |
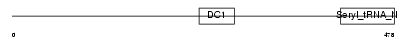
|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
KIF4A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.011432 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.005855 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.005966 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
ZFN2B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.011177 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WV99, Q8NB98 | Gene names | ZFAND2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 2B. | |||||
|
ZN561_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | -0.002141 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N587, Q6PJS0 | Gene names | ZNF561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 561. | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.069156 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.063530 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
MSH3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH3_HUMAN
|
||||||
| NC score | 0.992779 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH2_MOUSE
|
||||||
| NC score | 0.872697 (rank : 3) | θ value | 2.17557e-62 (rank : 8) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_HUMAN
|
||||||
| NC score | 0.872142 (rank : 4) | θ value | 4.38363e-63 (rank : 7) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH4_HUMAN
|
||||||
| NC score | 0.871615 (rank : 5) | θ value | 1.36269e-64 (rank : 6) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH4_MOUSE
|
||||||
| NC score | 0.870466 (rank : 6) | θ value | 7.98882e-65 (rank : 5) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.862072 (rank : 7) | θ value | 3.45727e-76 (rank : 4) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_HUMAN
|
||||||
| NC score | 0.855877 (rank : 8) | θ value | 1.65806e-78 (rank : 3) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH5_MOUSE
|
||||||
| NC score | 0.823997 (rank : 9) | θ value | 2.11701e-41 (rank : 9) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_HUMAN
|
||||||
| NC score | 0.822937 (rank : 10) | θ value | 6.15952e-41 (rank : 10) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.069156 (rank : 11) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.063530 (rank : 12) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
GDE5_MOUSE
|
||||||
| NC score | 0.040474 (rank : 13) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C0L9, Q3TLV6, Q80TD5, Q8BKJ7, Q8BKW7, Q8CFW2, Q9D759 | Gene names | Gde5, Kiaa1434, Prei4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative glycerophosphodiester phosphodiesterase 5 (EC 3.1.-.-) (Preimplantation protein 4). | |||||
|
GDE5_HUMAN
|
||||||
| NC score | 0.035050 (rank : 14) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NPB8, Q9BQL8, Q9NUX0 | Gene names | GDE5, KIAA1434 | |||
|
Domain Architecture |

|
|||||
| Description | Putative glycerophosphodiester phosphodiesterase 5 (EC 3.1.-.-). | |||||
|
ASGR1_MOUSE
|
||||||
| NC score | 0.024882 (rank : 15) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P34927, Q64363 | Gene names | Asgr1, Asgr-1 | |||
|
Domain Architecture |

|
|||||
| Description | Asialoglycoprotein receptor 1 (ASGPR 1) (ASGP-R 1) (Hepatic lectin 1) (MHL-1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.022741 (rank : 16) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.022158 (rank : 17) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
NIBA_HUMAN
|
||||||
| NC score | 0.021228 (rank : 18) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BZQ8, Q5TEM8, Q8TEI5, Q9H593, Q9H9Y8, Q9HCB9 | Gene names | C1orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Niban protein. | |||||
|
SAMH1_MOUSE
|
||||||
| NC score | 0.017075 (rank : 19) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q60710, Q91VK8 | Gene names | Samhd1, Mg11 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Interferon-gamma- inducible protein Mg11). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.016124 (rank : 20) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.015726 (rank : 21) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.015725 (rank : 22) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CT096_HUMAN
|
||||||
| NC score | 0.015682 (rank : 23) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NUD7, Q8N840, Q8NAX5 | Gene names | C20orf96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf96. | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.015506 (rank : 24) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.015446 (rank : 25) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.013759 (rank : 26) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.013459 (rank : 27) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.012961 (rank : 28) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYH14_MOUSE
|
||||||
| NC score | 0.012393 (rank : 29) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1225 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6URW6, Q80V64, Q80ZE6 | Gene names | Myh14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.012181 (rank : 30) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| NC score | 0.012118 (rank : 31) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.011740 (rank : 32) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.011545 (rank : 33) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
KIF4A_MOUSE
|
||||||
| NC score | 0.011432 (rank : 34) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1033 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P33174 | Gene names | Kif4a, Kif4, Kns4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
ZFN2B_HUMAN
|
||||||
| NC score | 0.011177 (rank : 35) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WV99, Q8NB98 | Gene names | ZFAND2B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AN1-type zinc finger protein 2B. | |||||
|
MYH14_HUMAN
|
||||||
| NC score | 0.011023 (rank : 36) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 995 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z406, Q5CZ75, Q6XYE4, Q76B62, Q8WV23, Q96I22, Q9BT27, Q9BW35, Q9H882 | Gene names | MYH14, KIAA2034 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-14 (Myosin heavy chain, nonmuscle IIc) (Nonmuscle myosin heavy chain IIc) (NMHC II-C). | |||||
|
TAC2N_MOUSE
|
||||||
| NC score | 0.010164 (rank : 37) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q91XT6, Q9D4A0 | Gene names | Mtac2d1 | |||
|
Domain Architecture |

|
|||||
| Description | Membrane targeting tandem C2 domain-containing protein 1 (Tandem C2 protein in nucleus) (Tac2-N). | |||||
|
DTNA_MOUSE
|
||||||
| NC score | 0.009446 (rank : 38) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D2N4, P97319, Q61498, Q61499, Q9QZZ5, Q9WUL9, Q9WUM0 | Gene names | Dtna, Dtn | |||
|
Domain Architecture |

|
|||||
| Description | Dystrobrevin alpha (Alpha-dystrobrevin). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.009136 (rank : 39) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
OLR1_HUMAN
|
||||||
| NC score | 0.008240 (rank : 40) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78380, Q2PP00, Q7Z484 | Gene names | OLR1, LOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) (hLOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.005966 (rank : 41) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.005855 (rank : 42) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ZN561_HUMAN
|
||||||
| NC score | -0.002141 (rank : 43) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 41 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N587, Q6PJS0 | Gene names | ZNF561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 561. | |||||