Please be patient as the page loads
|
MSH2_MOUSE
|
||||||
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MSH2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.994523 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH3_HUMAN
|
||||||
| θ value | 7.98882e-65 (rank : 3) | NC score | 0.871764 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH3_MOUSE
|
||||||
| θ value | 2.17557e-62 (rank : 4) | NC score | 0.872697 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH6_MOUSE
|
||||||
| θ value | 8.00737e-57 (rank : 5) | NC score | 0.831205 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 6) | NC score | 0.821127 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH4_HUMAN
|
||||||
| θ value | 2.49495e-50 (rank : 7) | NC score | 0.844972 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH4_MOUSE
|
||||||
| θ value | 8.87376e-48 (rank : 8) | NC score | 0.838642 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
MSH5_HUMAN
|
||||||
| θ value | 3.16345e-37 (rank : 9) | NC score | 0.805653 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_MOUSE
|
||||||
| θ value | 9.20422e-37 (rank : 10) | NC score | 0.805752 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
TBCD5_HUMAN
|
||||||
| θ value | 0.163984 (rank : 11) | NC score | 0.066459 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92609 | Gene names | TBC1D5, KIAA0210 | |||
|
Domain Architecture |
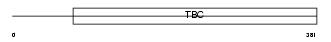
|
|||||
| Description | TBC1 domain family member 5. | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.038029 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.018368 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.038183 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 15) | NC score | 0.023230 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
TTL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.041781 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38585, Q8R1L7, Q8VEG2 | Gene names | Ttl | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
CP135_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.020153 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TTL_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.040608 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG68, Q7Z302, Q8N426 | Gene names | TTL | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
SUHW4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.012169 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
CF060_HUMAN
|
||||||
| θ value | 1.38821 (rank : 20) | NC score | 0.019562 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.017129 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
TBCD5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.052955 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XQ2 | Gene names | Tbc1d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 5. | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 23) | NC score | 0.016276 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.023382 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SUHW4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.011751 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
SUHW3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.016318 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
ING2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.024394 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H160, O95698 | Gene names | ING2, ING1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein) (ING1Lp) (p32). | |||||
|
CEP70_MOUSE
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.020780 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.022365 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DDX23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.007280 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
K2C3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.007103 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |
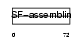
|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
HNRPD_HUMAN
|
||||||
| θ value | 5.27518 (rank : 32) | NC score | 0.007389 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |
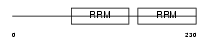
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.007404 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |
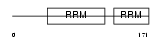
|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
MYH2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.016553 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.014193 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.016302 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
TJAP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.026261 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.014387 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
CD69_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.005172 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |
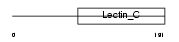
|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.019757 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
HAT1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.019903 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BY71 | Gene names | Hat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase type B catalytic subunit (EC 2.3.1.48). | |||||
|
ING2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.013199 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESK4, Q80VI5, Q8BGU8 | Gene names | Ing2, Ing1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein). | |||||
|
K0999_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | -0.000288 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P4S6, Q641L5, Q66JZ5, Q6ZQ09, Q8K075, Q9CYD5 | Gene names | Kiaa0999 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized serine/threonine-protein kinase KIAA0999 (EC 2.7.11.1). | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.010062 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
PAI2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.002035 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12388, O35687, Q9QWP6, Q9QWP7, Q9QWP8, Q9QWP9, Q9QWQ0, Q9QWZ5, Q9QWZ6 | Gene names | Serpinb2, Pai2, Planh2 | |||
|
Domain Architecture |
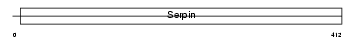
|
|||||
| Description | Plasminogen activator inhibitor 2, macrophage (PAI-2). | |||||
|
ZCPW1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.063422 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.057965 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
MSH2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P43247 | Gene names | Msh2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH2_HUMAN
|
||||||
| NC score | 0.994523 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P43246, O75488 | Gene names | MSH2 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh2 (MutS protein homolog 2). | |||||
|
MSH3_MOUSE
|
||||||
| NC score | 0.872697 (rank : 3) | θ value | 2.17557e-62 (rank : 4) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
MSH3_HUMAN
|
||||||
| NC score | 0.871764 (rank : 4) | θ value | 7.98882e-65 (rank : 3) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P20585, Q6PJT5, Q86UQ6, Q92867 | Gene names | MSH3, DUC1, DUG | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Divergent upstream protein) (DUP) (Mismatch repair protein 1) (MRP1). | |||||
|
MSH4_HUMAN
|
||||||
| NC score | 0.844972 (rank : 5) | θ value | 2.49495e-50 (rank : 7) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15457, Q8NEB3, Q9UNP8 | Gene names | MSH4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4. | |||||
|
MSH4_MOUSE
|
||||||
| NC score | 0.838642 (rank : 6) | θ value | 8.87376e-48 (rank : 8) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99MT2, Q920J1 | Gene names | Msh4 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 4 (mMsh4). | |||||
|
MSH6_MOUSE
|
||||||
| NC score | 0.831205 (rank : 7) | θ value | 8.00737e-57 (rank : 5) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P54276, O54710 | Gene names | Msh6, Gtmbp | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH6_HUMAN
|
||||||
| NC score | 0.821127 (rank : 8) | θ value | 3.36421e-55 (rank : 6) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52701, O43706, O43917, Q8TCX4, Q9BTB5 | Gene names | MSH6, GTBP | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein MSH6 (MutS-alpha 160 kDa subunit) (G/T mismatch-binding protein) (GTBP) (GTMBP) (p160). | |||||
|
MSH5_MOUSE
|
||||||
| NC score | 0.805752 (rank : 9) | θ value | 9.20422e-37 (rank : 10) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9QUM7, Q9Z1Q6 | Gene names | Msh5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MutS protein homolog 5. | |||||
|
MSH5_HUMAN
|
||||||
| NC score | 0.805653 (rank : 10) | θ value | 3.16345e-37 (rank : 9) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O43196, O60586, Q5BLU9, Q5SSR1, Q8IW44, Q9BQC7 | Gene names | MSH5 | |||
|
Domain Architecture |

|
|||||
| Description | MutS protein homolog 5. | |||||
|
TBCD5_HUMAN
|
||||||
| NC score | 0.066459 (rank : 11) | θ value | 0.163984 (rank : 11) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92609 | Gene names | TBC1D5, KIAA0210 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 5. | |||||
|
ZCPW1_HUMAN
|
||||||
| NC score | 0.063422 (rank : 12) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H0M4, Q8NA98, Q9BUD0, Q9NWF7 | Gene names | ZCWPW1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1. | |||||
|
ZCPW1_MOUSE
|
||||||
| NC score | 0.057965 (rank : 13) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6IR42 | Gene names | Zcwpw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CW-type PWWP domain protein 1 homolog. | |||||
|
TBCD5_MOUSE
|
||||||
| NC score | 0.052955 (rank : 14) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XQ2 | Gene names | Tbc1d5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TBC1 domain family member 5. | |||||
|
TTL_MOUSE
|
||||||
| NC score | 0.041781 (rank : 15) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P38585, Q8R1L7, Q8VEG2 | Gene names | Ttl | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
TTL_HUMAN
|
||||||
| NC score | 0.040608 (rank : 16) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NG68, Q7Z302, Q8N426 | Gene names | TTL | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase (EC 6.3.2.25) (TTL). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.038183 (rank : 17) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.038029 (rank : 18) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
TJAP1_MOUSE
|
||||||
| NC score | 0.026261 (rank : 19) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9DCD5, Q8CFL7, Q8R5I2 | Gene names | Tjap1, Pilt, Tjp4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
ING2_HUMAN
|
||||||
| NC score | 0.024394 (rank : 20) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H160, O95698 | Gene names | ING2, ING1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein) (ING1Lp) (p32). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.023382 (rank : 21) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.023230 (rank : 22) | θ value | 0.365318 (rank : 15) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.022365 (rank : 23) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CEP70_MOUSE
|
||||||
| NC score | 0.020780 (rank : 24) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.020153 (rank : 25) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
HAT1_MOUSE
|
||||||
| NC score | 0.019903 (rank : 26) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BY71 | Gene names | Hat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase type B catalytic subunit (EC 2.3.1.48). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.019757 (rank : 27) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.019562 (rank : 28) | θ value | 1.38821 (rank : 20) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.018368 (rank : 29) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.017129 (rank : 30) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH2_HUMAN
|
||||||
| NC score | 0.016553 (rank : 31) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1491 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UKX2, Q14322, Q16229 | Gene names | MYH2, MYHSA2 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, skeletal muscle, adult 2 (Myosin heavy chain IIa) (MyHC-IIa). | |||||
|
SUHW3_HUMAN
|
||||||
| NC score | 0.016318 (rank : 32) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8ND82, Q9NXR3 | Gene names | SUHW3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 3. | |||||
|
MYH1_HUMAN
|
||||||
| NC score | 0.016302 (rank : 33) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1478 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P12882, Q9Y622 | Gene names | MYH1 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1) (Myosin heavy chain IIx/d) (MyHC-IIx/d). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.016276 (rank : 34) | θ value | 1.81305 (rank : 23) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.014387 (rank : 35) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.014193 (rank : 36) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ING2_MOUSE
|
||||||
| NC score | 0.013199 (rank : 37) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESK4, Q80VI5, Q8BGU8 | Gene names | Ing2, Ing1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inhibitor of growth protein 2 (p33ING2) (Inhibitor of growth 1-like protein). | |||||
|
SUHW4_MOUSE
|
||||||
| NC score | 0.012169 (rank : 38) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 737 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q68FE8, Q3T9B1, Q8BI82 | Gene names | Suhw4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
SUHW4_HUMAN
|
||||||
| NC score | 0.011751 (rank : 39) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 860 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6N043, Q6MZM6, Q6N085, Q9H0U5, Q9HCI8, Q9NXS0 | Gene names | SUHW4, KIAA1584 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Suppressor of hairy wing homolog 4. | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.010062 (rank : 40) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
HNRPD_MOUSE
|
||||||
| NC score | 0.007404 (rank : 41) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60668, Q60667, Q80ZJ0, Q91X94 | Gene names | Hnrpd, Auf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
HNRPD_HUMAN
|
||||||
| NC score | 0.007389 (rank : 42) | θ value | 5.27518 (rank : 32) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14103, Q01858, Q14100, Q14101, Q14102 | Gene names | HNRPD, AUF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heterogeneous nuclear ribonucleoprotein D0 (hnRNP D0) (AU-rich element RNA-binding protein 1). | |||||
|
DDX23_HUMAN
|
||||||
| NC score | 0.007280 (rank : 43) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BUQ8, O43188 | Gene names | DDX23 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX23 (EC 3.6.1.-) (DEAD box protein 23) (100 kDa U5 snRNP-specific protein) (U5-100kD) (PRP28 homolog). | |||||
|
K2C3_HUMAN
|
||||||
| NC score | 0.007103 (rank : 44) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P12035, Q701L8 | Gene names | KRT3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 3 (Cytokeratin-3) (CK-3) (Keratin-3) (K3) (65 kDa cytokeratin). | |||||
|
CD69_HUMAN
|
||||||
| NC score | 0.005172 (rank : 45) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q07108 | Gene names | CD69 | |||
|
Domain Architecture |

|
|||||
| Description | Early activation antigen CD69 (Early T-cell activation antigen p60) (GP32/28) (Leu-23) (MLR-3) (EA1) (BL-AC/P26) (Activation inducer molecule) (AIM). | |||||
|
PAI2_MOUSE
|
||||||
| NC score | 0.002035 (rank : 46) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P12388, O35687, Q9QWP6, Q9QWP7, Q9QWP8, Q9QWP9, Q9QWQ0, Q9QWZ5, Q9QWZ6 | Gene names | Serpinb2, Pai2, Planh2 | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen activator inhibitor 2, macrophage (PAI-2). | |||||
|
K0999_MOUSE
|
||||||
| NC score | -0.000288 (rank : 47) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 45 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P4S6, Q641L5, Q66JZ5, Q6ZQ09, Q8K075, Q9CYD5 | Gene names | Kiaa0999 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized serine/threonine-protein kinase KIAA0999 (EC 2.7.11.1). | |||||