Please be patient as the page loads
|
EYA2_MOUSE
|
||||||
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EYA2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996046 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
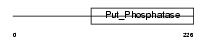
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA1_HUMAN
|
||||||
| θ value | 1.71888e-184 (rank : 3) | NC score | 0.975222 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| θ value | 5.01279e-176 (rank : 4) | NC score | 0.973625 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 3.84704e-168 (rank : 5) | NC score | 0.971330 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 2.76339e-158 (rank : 6) | NC score | 0.980370 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| θ value | 3.65119e-118 (rank : 7) | NC score | 0.978339 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| θ value | 4.03687e-117 (rank : 8) | NC score | 0.976439 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
HXB8_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.017619 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17481, Q9H1I2 | Gene names | HOXB8, HOX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2D) (Hox-2.4). | |||||
|
HXB8_MOUSE
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.017609 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09632 | Gene names | Hoxb8, Hox-2.4, Hoxb-8 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2.4). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.046657 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
FUS_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.108886 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
GPS2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.049842 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
P53_HUMAN
|
||||||
| θ value | 1.06291 (rank : 14) | NC score | 0.019876 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |
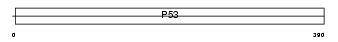
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
ZN575_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.001451 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.045187 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
DAZ1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.031099 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |
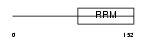
|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.031071 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
HXB3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.008109 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
IRX5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.010858 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |
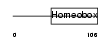
|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
FUS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.098355 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
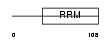
DAZ2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.029791 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
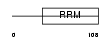
DAZ3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.028033 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |
|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
EST1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.011964 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
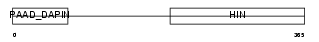
IFI3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.014531 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |
|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
LDB3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.014028 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
DEND_MOUSE
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.018038 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
FIGN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.005994 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
MSH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.012118 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
TBC12_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.008644 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
ZN438_HUMAN
|
||||||
| θ value | 5.27518 (rank : 31) | NC score | 0.001548 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
BAZ2A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.004947 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.018239 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
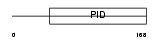
DAB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.008594 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |
|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
DBR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.010987 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK59, Q96GH0, Q9NXQ6 | Gene names | DBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lariat debranching enzyme (EC 3.1.-.-). | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.024069 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
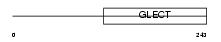
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.017510 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
CERU_HUMAN
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.005202 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
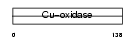
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
GLI3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | -0.000266 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||
|
MCM3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.035995 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.006213 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PDC6I_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.005966 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.007171 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
EWS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.094614 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
EWS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.097976 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
TFG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.051168 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
EYA2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.996046 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.980370 (rank : 3) | θ value | 2.76339e-158 (rank : 6) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA3_MOUSE
|
||||||
| NC score | 0.978339 (rank : 4) | θ value | 3.65119e-118 (rank : 7) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P97480, P97768 | Gene names | Eya3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA3_HUMAN
|
||||||
| NC score | 0.976439 (rank : 5) | θ value | 4.03687e-117 (rank : 8) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99504, O95463, Q99813 | Gene names | EYA3 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 3 (EC 3.1.3.48). | |||||
|
EYA1_HUMAN
|
||||||
| NC score | 0.975222 (rank : 6) | θ value | 1.71888e-184 (rank : 3) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99502 | Gene names | EYA1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA1_MOUSE
|
||||||
| NC score | 0.973625 (rank : 7) | θ value | 5.01279e-176 (rank : 4) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97767, O08818 | Gene names | Eya1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 1 (EC 3.1.3.48). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.971330 (rank : 8) | θ value | 3.84704e-168 (rank : 5) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.108886 (rank : 9) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.098355 (rank : 10) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.097976 (rank : 11) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.094614 (rank : 12) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
TFG_HUMAN
|
||||||
| NC score | 0.051168 (rank : 13) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
GPS2_HUMAN
|
||||||
| NC score | 0.049842 (rank : 14) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13227 | Gene names | GPS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G protein pathway suppressor 2 (Protein GPS2). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.046657 (rank : 15) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.045187 (rank : 16) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
MCM3A_HUMAN
|
||||||
| NC score | 0.035995 (rank : 17) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60318, Q9UMT4 | Gene names | MCM3AP, GANP, KIAA0572, MAP80 | |||
|
Domain Architecture |

|
|||||
| Description | 80 kDa MCM3-associated protein (GANP protein). | |||||
|
DAZ1_HUMAN
|
||||||
| NC score | 0.031099 (rank : 18) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NQZ3, Q9NQZ4 | Gene names | DAZ1, DAZ | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 1. | |||||
|
DAZ4_HUMAN
|
||||||
| NC score | 0.031071 (rank : 19) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q86SG3, Q9NR88, Q9NR89 | Gene names | DAZ4 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 4. | |||||
|
DAZ2_HUMAN
|
||||||
| NC score | 0.029791 (rank : 20) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13117, Q96P41, Q9NR91 | Gene names | DAZ2 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 2. | |||||
|
DAZ3_HUMAN
|
||||||
| NC score | 0.028033 (rank : 21) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR90 | Gene names | DAZ3 | |||
|
Domain Architecture |

|
|||||
| Description | Deleted in azoospermia protein 3. | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.024069 (rank : 22) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
P53_HUMAN
|
||||||
| NC score | 0.019876 (rank : 23) | θ value | 1.06291 (rank : 14) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.018239 (rank : 24) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
DEND_MOUSE
|
||||||
| NC score | 0.018038 (rank : 25) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80TS7 | Gene names | Ddn, Kiaa0749 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dendrin. | |||||
|
HXB8_HUMAN
|
||||||
| NC score | 0.017619 (rank : 26) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P17481, Q9H1I2 | Gene names | HOXB8, HOX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2D) (Hox-2.4). | |||||
|
HXB8_MOUSE
|
||||||
| NC score | 0.017609 (rank : 27) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09632 | Gene names | Hoxb8, Hox-2.4, Hoxb-8 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B8 (Hox-2.4). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.017510 (rank : 28) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
IFI3_MOUSE
|
||||||
| NC score | 0.014531 (rank : 29) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35368 | Gene names | Ifi203 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-activable protein 203 (Ifi-203) (Interferon-inducible protein p203). | |||||
|
LDB3_HUMAN
|
||||||
| NC score | 0.014028 (rank : 30) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75112, Q5K6N9, Q5K6P0, Q5K6P1, Q96FH2, Q9Y4Z3, Q9Y4Z4, Q9Y4Z5 | Gene names | LDB3, KIAA0613, ZASP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | LIM domain-binding protein 3 (Z-band alternatively spliced PDZ-motif protein) (Protein cypher). | |||||
|
MSH3_MOUSE
|
||||||
| NC score | 0.012118 (rank : 31) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P13705 | Gene names | Msh3, Rep-3 | |||
|
Domain Architecture |

|
|||||
| Description | DNA mismatch repair protein Msh3 (Repair-3 protein) (REP-1). | |||||
|
EST1A_MOUSE
|
||||||
| NC score | 0.011964 (rank : 32) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61406, Q5NC64 | Gene names | Est1a, Kiaa0732 | |||
|
Domain Architecture |

|
|||||
| Description | Telomerase-binding protein EST1A (Ever shorter telomeres 1A) (Telomerase subunit EST1A) (EST1-like protein A). | |||||
|
DBR1_HUMAN
|
||||||
| NC score | 0.010987 (rank : 33) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9UK59, Q96GH0, Q9NXQ6 | Gene names | DBR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lariat debranching enzyme (EC 3.1.-.-). | |||||
|
IRX5_HUMAN
|
||||||
| NC score | 0.010858 (rank : 34) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P78411, P78416, Q7Z2E1 | Gene names | IRX5, IRX2A, IRXB2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-5 (Iroquois homeobox protein 5) (Homeodomain protein IRXB2) (IRX-2A). | |||||
|
TBC12_HUMAN
|
||||||
| NC score | 0.008644 (rank : 35) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60347, Q5VYA6, Q8WX26, Q8WX59, Q9UG83 | Gene names | TBC1D12, KIAA0608 | |||
|
Domain Architecture |

|
|||||
| Description | TBC1 domain family member 12. | |||||
|
DAB2_MOUSE
|
||||||
| NC score | 0.008594 (rank : 36) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98078, Q3U3K1, Q91W56, Q923E1 | Gene names | Dab2, Doc2 | |||
|
Domain Architecture |

|
|||||
| Description | Disabled homolog 2 (DOC-2) (Mitogen-responsive phosphoprotein). | |||||
|
HXB3_MOUSE
|
||||||
| NC score | 0.008109 (rank : 37) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09026, P10285, Q4PJ06, Q61680 | Gene names | Hoxb3, Hox-2.7, Hoxb-3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B3 (Hox-2.7) (Homeobox protein MH-23). | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.007171 (rank : 38) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.006213 (rank : 39) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
FIGN_HUMAN
|
||||||
| NC score | 0.005994 (rank : 40) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5HY92, Q9H6M5, Q9NVZ9 | Gene names | FIGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fidgetin. | |||||
|
PDC6I_MOUSE
|
||||||
| NC score | 0.005966 (rank : 41) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WU78, O88695, O89014, Q8BSL8, Q8R0H5, Q99LR3, Q9QZN8 | Gene names | Pdcd6ip, Aip1, Alix | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (ALG-2-interacting protein X) (ALG-2-interacting protein 1) (E2F1-inducible protein) (Eig2). | |||||
|
CERU_HUMAN
|
||||||
| NC score | 0.005202 (rank : 42) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00450, Q14063, Q2PP18, Q9UKS4 | Gene names | CP | |||
|
Domain Architecture |
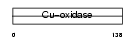
|
|||||
| Description | Ceruloplasmin precursor (EC 1.16.3.1) (Ferroxidase). | |||||
|
BAZ2A_HUMAN
|
||||||
| NC score | 0.004947 (rank : 43) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UIF9, O00536, O15030, Q96H26 | Gene names | BAZ2A, KIAA0314, TIP5 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain 2A (Transcription termination factor I-interacting protein 5) (TTF-I-interacting protein 5) (Tip5) (hWALp3). | |||||
|
ZN438_HUMAN
|
||||||
| NC score | 0.001548 (rank : 44) | θ value | 5.27518 (rank : 31) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z4V0, Q5T426, Q658Q4, Q6ZN65 | Gene names | ZNF438 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 438. | |||||
|
ZN575_HUMAN
|
||||||
| NC score | 0.001451 (rank : 45) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86XF7 | Gene names | ZNF575 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 575. | |||||
|
GLI3_MOUSE
|
||||||
| NC score | -0.000266 (rank : 46) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 43 | Target Neighborhood Hits | 807 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61602 | Gene names | Gli3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI3. | |||||