Please be patient as the page loads
|
P53_HUMAN
|
||||||
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |
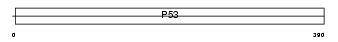
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P53_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
P53_MOUSE
|
||||||
| θ value | 9.80574e-164 (rank : 2) | NC score | 0.989168 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |
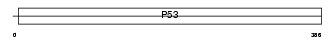
|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
|
P73L_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 3) | NC score | 0.905679 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
P73L_MOUSE
|
||||||
| θ value | 4.67263e-73 (rank : 4) | NC score | 0.907222 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73_HUMAN
|
||||||
| θ value | 3.95564e-72 (rank : 5) | NC score | 0.919054 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
GNAS1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.023940 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.053314 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
PDLI7_HUMAN
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.022968 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.021359 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
AKAP5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.079215 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |
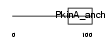
|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
RIF1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.031407 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
EYA2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.019876 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
STON2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.025919 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
K1802_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.021385 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.044112 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
MKL1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 16) | NC score | 0.020027 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
K1802_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.021412 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
LIMD1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.015415 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MINT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.014888 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MKRN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 20) | NC score | 0.015727 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
DNM3A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.014250 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.020443 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.020543 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
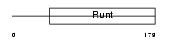
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TOPB1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.020261 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
G3BP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.012607 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.004644 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
ZFP28_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | -0.000314 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |
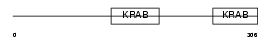
|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.010728 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
C1QR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.003725 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |
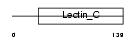
|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
CSF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.014481 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
HLX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.004286 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61670 | Gene names | Hlx1, Hlx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.008232 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.000496 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.007397 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
3BP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.011414 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CF155_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.023725 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.012678 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
GLIS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 38) | NC score | 0.000242 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.005416 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.005732 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
THOC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.011515 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
UBQL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | 0.008921 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
EVI2B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.068080 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
P53_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P04637, Q15086, Q15087, Q15088, Q16535, Q16807, Q16808, Q16809, Q16810, Q16811, Q16848, Q86UG1, Q8J016, Q99659, Q9BTM4, Q9HAQ8, Q9NP68, Q9NPJ2, Q9NZD0, Q9UBI2, Q9UQ61 | Gene names | TP53, P53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53) (Phosphoprotein p53) (Antigen NY-CO-13). | |||||
|
P53_MOUSE
|
||||||
| NC score | 0.989168 (rank : 2) | θ value | 9.80574e-164 (rank : 2) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P02340, Q9QUP3 | Gene names | Tp53, P53, Trp53 | |||
|
Domain Architecture |

|
|||||
| Description | Cellular tumor antigen p53 (Tumor suppressor p53). | |||||
|
P73_HUMAN
|
||||||
| NC score | 0.919054 (rank : 3) | θ value | 3.95564e-72 (rank : 5) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O15350, O15351, Q5TBV5, Q5TBV6, Q8NHW9, Q8TDY5, Q8TDY6, Q9NTK8 | Gene names | TP73, P73 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein p73 (p53-like transcription factor) (p53-related protein). | |||||
|
P73L_MOUSE
|
||||||
| NC score | 0.907222 (rank : 4) | θ value | 4.67263e-73 (rank : 4) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88898, O88897, O88899, O89097, Q8C826, Q9QWY9, Q9QWZ0 | Gene names | Tp73l, P63, P73l, Trp63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Transformation-related protein 63). | |||||
|
P73L_HUMAN
|
||||||
| NC score | 0.905679 (rank : 5) | θ value | 4.67263e-73 (rank : 3) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H3D4, O75080, O75195, O75922, O76078, Q6VEG2, Q6VEG3, Q6VEG4, Q6VFJ1, Q6VFJ2, Q6VFJ3, Q6VH20, Q7LDI3, Q7LDI4, Q7LDI5, Q96KR0, Q9H3D2, Q9H3D3, Q9H3P8, Q9NPH7, Q9P1B4, Q9P1B5, Q9P1B6, Q9P1B7, Q9UBV9, Q9UE10, Q9UP26, Q9UP27, Q9UP28, Q9UP74 | Gene names | TP73L, KET, P63, P73H, P73L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tumor protein p73-like (p73L) (p63) (Tumor protein 63) (TP63) (p51) (p40) (Keratinocyte transcription factor KET) (Chronic ulcerative stomatitis protein) (CUSP). | |||||
|
AKAP5_HUMAN
|
||||||
| NC score | 0.079215 (rank : 6) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P24588 | Gene names | AKAP5, AKAP79 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 5 (A-kinase anchor protein 79 kDa) (AKAP 79) (cAMP-dependent protein kinase regulatory subunit II high affinity- binding protein) (H21). | |||||
|
EVI2B_MOUSE
|
||||||
| NC score | 0.068080 (rank : 7) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.053314 (rank : 8) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.044112 (rank : 9) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
RIF1_HUMAN
|
||||||
| NC score | 0.031407 (rank : 10) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 320 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5UIP0, Q5H9R3, Q5UIP2, Q66YK6, Q6PRU2, Q8TE94, Q99772, Q9H830, Q9H9B9, Q9NVP5, Q9Y4R4 | Gene names | RIF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Telomere-associated protein RIF1 (Rap1-interacting factor 1 homolog). | |||||
|
STON2_MOUSE
|
||||||
| NC score | 0.025919 (rank : 11) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BZ60 | Gene names | Ston2, Stn2, Stnb | |||
|
Domain Architecture |

|
|||||
| Description | Stonin-2 (Stoned B). | |||||
|
GNAS1_MOUSE
|
||||||
| NC score | 0.023940 (rank : 12) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 651 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6R0H7, Q6R0H4, Q6R0H5, Q6R2J5, Q9JJX0, Q9Z1N8 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanine nucleotide-binding protein G(s) subunit alpha isoforms XLas (Adenylate cyclase-stimulating G alpha protein) (Extra large alphas protein) (XLalphas). | |||||
|
CF155_HUMAN
|
||||||
| NC score | 0.023725 (rank : 13) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8W2, Q5TG13 | Gene names | C6orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C6orf155. | |||||
|
PDLI7_HUMAN
|
||||||
| NC score | 0.022968 (rank : 14) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 547 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9NR12, Q14250, Q5XG82, Q6NVZ5, Q96C91, Q9BXB8, Q9BXB9 | Gene names | PDLIM7, ENIGMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PDZ and LIM domain protein 7 (LIM mineralization protein) (LMP) (Protein enigma). | |||||
|
K1802_HUMAN
|
||||||
| NC score | 0.021412 (rank : 15) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1378 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96JM3, Q6P181, Q8NC88, Q9BST0 | Gene names | KIAA1802, C13orf8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
K1802_MOUSE
|
||||||
| NC score | 0.021385 (rank : 16) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1596 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8K327, Q3UZ85, Q6ZPI1 | Gene names | Kiaa1802, D8Ertd457e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein KIAA1802. | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.021359 (rank : 17) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.020543 (rank : 18) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.020443 (rank : 19) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
TOPB1_MOUSE
|
||||||
| NC score | 0.020261 (rank : 20) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZQF0, Q6P6P0, Q80Y33, Q8BUI0, Q8BUK1, Q8R348, Q91VX3, Q922X8 | Gene names | Topbp1, Kiaa0259 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA topoisomerase II-binding protein 1 (DNA topoisomerase IIbeta- binding protein 1) (TopBP1). | |||||
|
MKL1_MOUSE
|
||||||
| NC score | 0.020027 (rank : 21) | θ value | 1.81305 (rank : 16) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 667 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4J6 | Gene names | Mkl1, Bsac | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein homolog) (Basic SAP coiled-coil transcription activator). | |||||
|
EYA2_MOUSE
|
||||||
| NC score | 0.019876 (rank : 22) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08575, P97925 | Gene names | Eya2, Eab1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
MKRN3_HUMAN
|
||||||
| NC score | 0.015727 (rank : 23) | θ value | 3.0926 (rank : 20) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13064 | Gene names | MKRN3, RNF63, ZNF127 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-3 (Zinc finger protein 127) (RING finger protein 63). | |||||
|
LIMD1_MOUSE
|
||||||
| NC score | 0.015415 (rank : 24) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QXD8, Q8C8G4, Q9CW55 | Gene names | Limd1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM domain-containing protein 1. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.014888 (rank : 25) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CSF1_HUMAN
|
||||||
| NC score | 0.014481 (rank : 26) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09603, Q13130, Q14086, Q14806, Q9UQR8 | Gene names | CSF1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage colony-stimulating factor 1 precursor (CSF-1) (MCSF) (M- CSF) (Lanimostim). | |||||
|
DNM3A_HUMAN
|
||||||
| NC score | 0.014250 (rank : 27) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K1, Q8WXU9 | Gene names | DNMT3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 3A (EC 2.1.1.37) (Dnmt3a) (DNA methyltransferase HsaIIIA) (DNA MTase HsaIIIA) (M.HsaIIIA). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.012678 (rank : 28) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
G3BP_HUMAN
|
||||||
| NC score | 0.012607 (rank : 29) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13283 | Gene names | G3BP, G3BP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-GTPase-activating protein-binding protein 1 (EC 3.6.1.-) (ATP- dependent DNA helicase VIII) (GAP SH3-domain-binding protein 1) (G3BP- 1) (HDH-VIII). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.011515 (rank : 30) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
3BP2_HUMAN
|
||||||
| NC score | 0.011414 (rank : 31) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P78314, O00500, O15373, P78315 | Gene names | SH3BP2, 3BP2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.010728 (rank : 32) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
UBQL3_HUMAN
|
||||||
| NC score | 0.008921 (rank : 33) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H347, Q9NRE0 | Gene names | UBQLN3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.008232 (rank : 34) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.007397 (rank : 35) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.005732 (rank : 36) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.005416 (rank : 37) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.004644 (rank : 38) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
HLX1_MOUSE
|
||||||
| NC score | 0.004286 (rank : 39) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61670 | Gene names | Hlx1, Hlx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein HLX1. | |||||
|
C1QR1_HUMAN
|
||||||
| NC score | 0.003725 (rank : 40) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 493 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NPY3, O00274 | Gene names | CD93, C1QR1, MXRA4 | |||
|
Domain Architecture |

|
|||||
| Description | Complement component C1q receptor precursor (Complement component 1 q subcomponent receptor 1) (C1qR) (C1qRp) (C1qR(p)) (C1q/MBL/SPA receptor) (CD93 antigen) (CDw93). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.000496 (rank : 41) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
GLIS3_MOUSE
|
||||||
| NC score | 0.000242 (rank : 42) | θ value | 8.99809 (rank : 38) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 861 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6XP49, Q8BI60 | Gene names | Glis3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLIS3 (GLI-similar 3). | |||||
|
ZFP28_HUMAN
|
||||||
| NC score | -0.000314 (rank : 43) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 42 | Target Neighborhood Hits | 843 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NHY6, Q9BY30, Q9P2B6 | Gene names | ZFP28, KIAA1431 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 28 homolog (Zfp-28) (Krueppel-like zinc finger factor X6). | |||||