Please be patient as the page loads
|
RUNX3_MOUSE
|
||||||
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
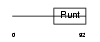
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RUNX3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.978505 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |
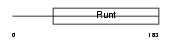
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 2.87963e-131 (rank : 3) | NC score | 0.953450 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 1.33764e-128 (rank : 4) | NC score | 0.955203 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
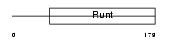
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 6.45994e-107 (rank : 5) | NC score | 0.943304 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 3.54468e-105 (rank : 6) | NC score | 0.938492 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.0148317 (rank : 7) | NC score | 0.052896 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SYNPO_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 8) | NC score | 0.065794 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 9) | NC score | 0.078137 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 10) | NC score | 0.098327 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 11) | NC score | 0.019962 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.019969 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 13) | NC score | 0.076929 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 14) | NC score | 0.033229 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
EP400_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 15) | NC score | 0.039122 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 16) | NC score | 0.051550 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.068295 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.054103 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.050457 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
HXD9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.017123 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28356 | Gene names | HOXD9, HOX4C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D9 (Hox-4C) (Hox-5.2). | |||||
|
NANO1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.046498 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WY41 | Gene names | NANOS1, NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1 (NOS-1) (EC_Rep1a). | |||||
|
ADNP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.036360 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
CHCH2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.077313 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6H1, Q6NZ50 | Gene names | CHCHD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2 (HCV NS2 trans-regulated protein) (NS2TP). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.057049 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.046752 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
IRS2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.047027 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.053605 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ATS13_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.011197 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.037804 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
TJAP1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.027288 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
AUTS2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.036266 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.037528 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.081790 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
EGR4_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.003960 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.016269 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.015909 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
PO3F3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.019531 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31361 | Gene names | Pou3f3, Brn-1, Brn1, Otf8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.062764 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.063666 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
TM108_HUMAN
|
||||||
| θ value | 1.06291 (rank : 40) | NC score | 0.044853 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CITE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.029325 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99966 | Gene names | CITED1, MSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.045782 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
IDD_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.024462 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |
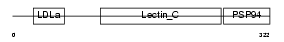
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.039284 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
PO3F3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.019111 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20264, P78379 | Gene names | POU3F3, BRN1, OTF8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
ZN703_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.038814 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7S9, Q5XG76 | Gene names | ZNF703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 703. | |||||
|
CHCH2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.057097 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1L0, Q3TUG8 | Gene names | Chchd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.041346 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.050939 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CNOT4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.036431 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
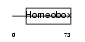
HXA10_HUMAN
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.013169 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31260, O43370, O43605, Q15949 | Gene names | HOXA10, HOX1H | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein Hox-A10 (Hox-1H) (Hox-1.8) (PL). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 52) | NC score | 0.046777 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
SC24B_HUMAN
|
||||||
| θ value | 1.81305 (rank : 53) | NC score | 0.020226 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
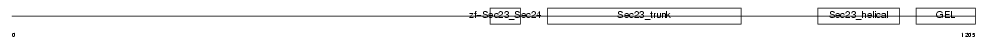
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
SGIP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 54) | NC score | 0.031745 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 55) | NC score | 0.063954 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
TM108_MOUSE
|
||||||
| θ value | 1.81305 (rank : 56) | NC score | 0.048672 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ARX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 57) | NC score | 0.013130 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
ARX_MOUSE
|
||||||
| θ value | 2.36792 (rank : 58) | NC score | 0.013514 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35085, Q9QYT4 | Gene names | Arx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
ASTL_HUMAN
|
||||||
| θ value | 2.36792 (rank : 59) | NC score | 0.024420 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
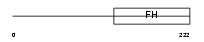
FOXD1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 60) | NC score | 0.006548 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
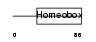
GBX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 61) | NC score | 0.014487 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52951, O43833, Q53RX5, Q9Y5Y1 | Gene names | GBX2 | |||
|
Domain Architecture |
|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2). | |||||
|
GBX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 62) | NC score | 0.014511 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48031 | Gene names | Gbx2, Mmoxa, Stra7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2) (Homeobox protein STRA7). | |||||
|
IRS1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 63) | NC score | 0.034774 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
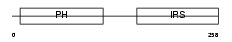
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
SP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 64) | NC score | 0.003728 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.034306 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
ASH2L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.018506 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.025641 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.035611 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.033577 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
MAFB_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.018917 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 3.0926 (rank : 71) | NC score | 0.021830 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
SP5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 72) | NC score | 0.003599 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 73) | NC score | 0.070867 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TBX10_MOUSE
|
||||||
| θ value | 3.0926 (rank : 74) | NC score | 0.007536 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810F8, O54841, Q6QH75 | Gene names | Tbx10, Tbx13, Tbx7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10) (T-box protein 13) (MmTBX7). | |||||
|
CCDC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.027022 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
MAFB_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.018029 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.033303 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
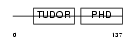
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.008079 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.024468 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TS101_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.024128 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |
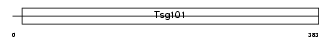
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.045928 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CDON_MOUSE
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.002790 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
CDX2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.010934 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99626, O00503, Q969L8 | Gene names | CDX2, CDX3 | |||
|
Domain Architecture |
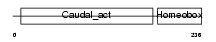
|
|||||
| Description | Homeobox protein CDX-2 (Caudal-type homeobox protein 2) (CDX-3). | |||||
|
DACH1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.013683 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
DBP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.011961 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
EYA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.008330 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |
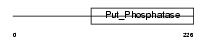
|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
NCOA5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.017815 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.024166 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.034183 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.037785 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 91) | NC score | 0.012509 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
EGR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 92) | NC score | 0.003923 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
GLI1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 93) | NC score | 0.003053 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.029721 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.020422 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
NPTXR_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.007868 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
PI5PA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.039550 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
RBM14_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.022621 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
RBM14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.021448 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 100) | NC score | 0.016319 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
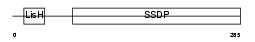
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 101) | NC score | 0.016319 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
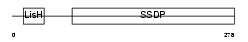
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.024236 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
DDX54_HUMAN
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.002192 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||
|
ENAH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.018423 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
FOXD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.004775 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |
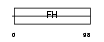
|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
IL3B2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | 0.014020 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
KBTBB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.005541 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94819 | Gene names | KBTBD11, KIAA0711, KLHDC7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch repeat and BTB domain-containing protein 11 (Kelch domain- containing protein 7B). | |||||
|
NRG3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.023591 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56975 | Gene names | NRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.025830 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.017647 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TAF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.021448 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
WASF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.009637 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
ZBTB4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.002799 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_HUMAN
|
||||||
| NC score | 0.978505 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.955203 (rank : 3) | θ value | 1.33764e-128 (rank : 4) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.953450 (rank : 4) | θ value | 2.87963e-131 (rank : 3) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.943304 (rank : 5) | θ value | 6.45994e-107 (rank : 5) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.938492 (rank : 6) | θ value | 3.54468e-105 (rank : 6) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.098327 (rank : 7) | θ value | 0.0252991 (rank : 10) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.081790 (rank : 8) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.078137 (rank : 9) | θ value | 0.0252991 (rank : 9) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CHCH2_HUMAN
|
||||||
| NC score | 0.077313 (rank : 10) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y6H1, Q6NZ50 | Gene names | CHCHD2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2 (HCV NS2 trans-regulated protein) (NS2TP). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.076929 (rank : 11) | θ value | 0.0563607 (rank : 13) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.070867 (rank : 12) | θ value | 3.0926 (rank : 73) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.068295 (rank : 13) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SYNPO_HUMAN
|
||||||
| NC score | 0.065794 (rank : 14) | θ value | 0.0148317 (rank : 8) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 281 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8N3V7, O15271, Q9UPX1 | Gene names | SYNPO, KIAA1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.063954 (rank : 15) | θ value | 1.81305 (rank : 55) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.063666 (rank : 16) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.062764 (rank : 17) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CHCH2_MOUSE
|
||||||
| NC score | 0.057097 (rank : 18) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D1L0, Q3TUG8 | Gene names | Chchd2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2. | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.057049 (rank : 19) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.054103 (rank : 20) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.053605 (rank : 21) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.052896 (rank : 22) | θ value | 0.0148317 (rank : 7) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.051550 (rank : 23) | θ value | 0.0961366 (rank : 16) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.050939 (rank : 24) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.050457 (rank : 25) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.048672 (rank : 26) | θ value | 1.81305 (rank : 56) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
IRS2_HUMAN
|
||||||
| NC score | 0.047027 (rank : 27) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y4H2, Q96RR2, Q9BZG0, Q9Y6I5 | Gene names | IRS2 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 2 (IRS-2). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.046777 (rank : 28) | θ value | 1.81305 (rank : 52) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.046752 (rank : 29) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
NANO1_HUMAN
|
||||||
| NC score | 0.046498 (rank : 30) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WY41 | Gene names | NANOS1, NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1 (NOS-1) (EC_Rep1a). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.045928 (rank : 31) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.045782 (rank : 32) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.044853 (rank : 33) | θ value | 1.06291 (rank : 40) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.041346 (rank : 34) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
PI5PA_MOUSE
|
||||||
| NC score | 0.039550 (rank : 35) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59644 | Gene names | Pib5pa | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.039284 (rank : 36) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.039122 (rank : 37) | θ value | 0.0961366 (rank : 15) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
ZN703_HUMAN
|
||||||
| NC score | 0.038814 (rank : 38) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H7S9, Q5XG76 | Gene names | ZNF703 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 703. | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.037804 (rank : 39) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.037785 (rank : 40) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.037528 (rank : 41) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CNOT4_HUMAN
|
||||||
| NC score | 0.036431 (rank : 42) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95628, O95339, O95627, Q8IYM7, Q8NCL0, Q9NPQ1, Q9NZN6 | Gene names | CNOT4, NOT4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
ADNP_HUMAN
|
||||||
| NC score | 0.036360 (rank : 43) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H2P0, O94881, Q9UG34 | Gene names | ADNP, KIAA0784 | |||
|
Domain Architecture |

|
|||||
| Description | Activity-dependent neuroprotector (Activity-dependent neuroprotective protein). | |||||
|
AUTS2_HUMAN
|
||||||
| NC score | 0.036266 (rank : 44) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8WXX7, Q9Y4F2 | Gene names | AUTS2, KIAA0442 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Autism susceptibility gene 2 protein. | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.035611 (rank : 45) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.034774 (rank : 46) | θ value | 2.36792 (rank : 63) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.034306 (rank : 47) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.034183 (rank : 48) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.033577 (rank : 49) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.033303 (rank : 50) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.033229 (rank : 51) | θ value | 0.0736092 (rank : 14) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
SGIP1_MOUSE
|
||||||
| NC score | 0.031745 (rank : 52) | θ value | 1.81305 (rank : 54) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VD37, Q3UFU3, Q3UGA0, Q8BXX4, Q8C034, Q9CXT2 | Gene names | Sgip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3-containing GRB2-like protein 3-interacting protein 1 (Endophilin- 3-interacting protein). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.029721 (rank : 53) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
CITE1_HUMAN
|
||||||
| NC score | 0.029325 (rank : 54) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99966 | Gene names | CITED1, MSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Cbp/p300-interacting transactivator 1 (Melanocyte-specific protein 1). | |||||
|
TJAP1_HUMAN
|
||||||
| NC score | 0.027288 (rank : 55) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q5JTD0, Q5JTD1, Q5JWW1, Q68DB2, Q6P2P3, Q9H7V7 | Gene names | TJAP1, PILT, TJP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tight junction-associated protein 1 (Tight junction protein 4) (Protein incorporated later into tight junctions). | |||||
|
CCDC4_HUMAN
|
||||||
| NC score | 0.027022 (rank : 56) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZU67, Q58A26, Q58A27 | Gene names | CCDC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 4. | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.025830 (rank : 57) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.025641 (rank : 58) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.024468 (rank : 59) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
IDD_HUMAN
|
||||||
| NC score | 0.024462 (rank : 60) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
ASTL_HUMAN
|
||||||
| NC score | 0.024420 (rank : 61) | θ value | 2.36792 (rank : 59) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6HA08 | Gene names | ASTL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Astacin-like metalloendopeptidase precursor (EC 3.4.-.-) (Oocyte astacin) (Ovastacin). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.024236 (rank : 62) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.024166 (rank : 63) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
TS101_MOUSE
|
||||||
| NC score | 0.024128 (rank : 64) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
NRG3_HUMAN
|
||||||
| NC score | 0.023591 (rank : 65) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56975 | Gene names | NRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
RBM14_HUMAN
|
||||||
| NC score | 0.022621 (rank : 66) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96PK6, O75932, Q53GV1, Q68DQ9, Q96PK5 | Gene names | RBM14, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14) (RRM-containing coactivator activator/modulator) (Synaptotagmin-interacting protein) (SYT-interacting protein). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.021830 (rank : 67) | θ value | 3.0926 (rank : 71) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.021448 (rank : 68) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
RBM14_MOUSE
|
||||||
| NC score | 0.021448 (rank : 69) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C2Q3, Q3TJB6, Q91Z21, Q9DBI6 | Gene names | Rbm14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA-binding protein 14 (RNA-binding motif protein 14). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.020422 (rank : 70) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SC24B_HUMAN
|
||||||
| NC score | 0.020226 (rank : 71) | θ value | 1.81305 (rank : 53) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95487 | Gene names | SEC24B | |||
|
Domain Architecture |
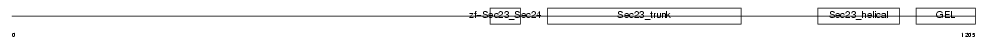
|
|||||
| Description | Protein transport protein Sec24B (SEC24-related protein B). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.019969 (rank : 72) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.019962 (rank : 73) | θ value | 0.0330416 (rank : 11) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
PO3F3_MOUSE
|
||||||
| NC score | 0.019531 (rank : 74) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P31361 | Gene names | Pou3f3, Brn-1, Brn1, Otf8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
PO3F3_HUMAN
|
||||||
| NC score | 0.019111 (rank : 75) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P20264, P78379 | Gene names | POU3F3, BRN1, OTF8 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 3, transcription factor 3 (Brain-specific homeobox/POU domain protein 1) (Brain-1) (Brn-1 protein). | |||||
|
MAFB_HUMAN
|
||||||
| NC score | 0.018917 (rank : 76) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Q3, Q9H1F1 | Gene names | MAFB, KRML | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B). | |||||
|
ASH2L_HUMAN
|
||||||
| NC score | 0.018506 (rank : 77) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBL3, O60659, O60660, Q96B62 | Gene names | ASH2L | |||
|
Domain Architecture |

|
|||||
| Description | Set1/Ash2 histone methyltransferase complex subunit ASH2 (ASH2-like protein). | |||||
|
ENAH_HUMAN
|
||||||
| NC score | 0.018423 (rank : 78) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8N8S7, Q502W5, Q5T5M7, Q5VTQ9, Q5VTR0, Q9NVF3, Q9UFB8 | Gene names | ENAH, MENA | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog. | |||||
|
MAFB_MOUSE
|
||||||
| NC score | 0.018029 (rank : 79) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54841 | Gene names | Mafb, Krml, Maf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor MafB (V-maf musculoaponeurotic fibrosarcoma oncogene homolog B) (Transcription factor MAF1) (Segmentation protein KR) (Kreisler). | |||||
|
NCOA5_HUMAN
|
||||||
| NC score | 0.017815 (rank : 80) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HCD5, Q6HA99, Q9H1F2, Q9H2T2, Q9H4Y9 | Gene names | NCOA5, KIAA1637 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor coactivator 5 (NCoA-5) (Coactivator independent of AF-2) (CIA). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.017647 (rank : 81) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
HXD9_HUMAN
|
||||||
| NC score | 0.017123 (rank : 82) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P28356 | Gene names | HOXD9, HOX4C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D9 (Hox-4C) (Hox-5.2). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.016319 (rank : 83) | θ value | 6.88961 (rank : 100) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.016319 (rank : 84) | θ value | 6.88961 (rank : 101) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.016269 (rank : 85) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.015909 (rank : 86) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
GBX2_MOUSE
|
||||||
| NC score | 0.014511 (rank : 87) | θ value | 2.36792 (rank : 62) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P48031 | Gene names | Gbx2, Mmoxa, Stra7 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2) (Homeobox protein STRA7). | |||||
|
GBX2_HUMAN
|
||||||
| NC score | 0.014487 (rank : 88) | θ value | 2.36792 (rank : 61) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P52951, O43833, Q53RX5, Q9Y5Y1 | Gene names | GBX2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GBX-2 (Gastrulation and brain-specific homeobox protein 2). | |||||
|
IL3B2_MOUSE
|
||||||
| NC score | 0.014020 (rank : 89) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 372 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P26954 | Gene names | Csf2rb2, Ai2ca, Il3r, Il3rb2 | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-3 receptor class 2 beta chain precursor (Interleukin-3 receptor class II beta chain) (Colony-stimulating factor 2 receptor, beta 2 chain). | |||||
|
DACH1_MOUSE
|
||||||
| NC score | 0.013683 (rank : 90) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QYB2, O88716, Q8BPQ0, Q8C8D7, Q9QY47, Q9R218, Q9Z0Y5 | Gene names | Dach1, Dach | |||
|
Domain Architecture |

|
|||||
| Description | Dachshund homolog 1 (Dach1). | |||||
|
ARX_MOUSE
|
||||||
| NC score | 0.013514 (rank : 91) | θ value | 2.36792 (rank : 58) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O35085, Q9QYT4 | Gene names | Arx | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
HXA10_HUMAN
|
||||||
| NC score | 0.013169 (rank : 92) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P31260, O43370, O43605, Q15949 | Gene names | HOXA10, HOX1H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A10 (Hox-1H) (Hox-1.8) (PL). | |||||
|
ARX_HUMAN
|
||||||
| NC score | 0.013130 (rank : 93) | θ value | 2.36792 (rank : 57) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96QS3 | Gene names | ARX | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein ARX (Aristaless-related homeobox). | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.012509 (rank : 94) | θ value | 6.88961 (rank : 91) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.011961 (rank : 95) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
ATS13_MOUSE
|
||||||
| NC score | 0.011197 (rank : 96) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q769J6, Q76LW1 | Gene names | Adamts13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ADAMTS-13 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 13) (ADAM-TS 13) (ADAM-TS13) (von Willebrand factor-cleaving protease) (vWF-cleaving protease) (vWF-CP). | |||||
|
CDX2_HUMAN
|
||||||
| NC score | 0.010934 (rank : 97) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99626, O00503, Q969L8 | Gene names | CDX2, CDX3 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-2 (Caudal-type homeobox protein 2) (CDX-3). | |||||
|
WASF3_MOUSE
|
||||||
| NC score | 0.009637 (rank : 98) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8VHI6 | Gene names | Wasf3, Wave3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein family member 3 (WASP-family protein member 3) (Protein WAVE-3). | |||||
|
EYA2_HUMAN
|
||||||
| NC score | 0.008330 (rank : 99) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O00167, Q5JSW8, Q96CV6, Q96H97, Q99503, Q99812, Q9BWF6, Q9H4S3, Q9H4S9, Q9NPZ4, Q9UIX7 | Gene names | EYA2, EAB1 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 2 (EC 3.1.3.48). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.008079 (rank : 100) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
NPTXR_HUMAN
|
||||||
| NC score | 0.007868 (rank : 101) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
TBX10_MOUSE
|
||||||
| NC score | 0.007536 (rank : 102) | θ value | 3.0926 (rank : 74) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810F8, O54841, Q6QH75 | Gene names | Tbx10, Tbx13, Tbx7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10) (T-box protein 13) (MmTBX7). | |||||
|
FOXD1_MOUSE
|
||||||
| NC score | 0.006548 (rank : 103) | θ value | 2.36792 (rank : 60) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61345 | Gene names | Foxd1, Fkhl8, Freac4, Hfhbf2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4) (Brain factor 2) (HFH-BF-2). | |||||
|
KBTBB_HUMAN
|
||||||
| NC score | 0.005541 (rank : 104) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O94819 | Gene names | KBTBD11, KIAA0711, KLHDC7C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kelch repeat and BTB domain-containing protein 11 (Kelch domain- containing protein 7B). | |||||
|
FOXD1_HUMAN
|
||||||
| NC score | 0.004775 (rank : 105) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16676, Q12949 | Gene names | FOXD1, FKHL8, FREAC4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein D1 (Forkhead-related protein FKHL8) (Forkhead- related transcription factor 4) (FREAC-4). | |||||
|
EGR4_MOUSE
|
||||||
| NC score | 0.003960 (rank : 106) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 710 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WUF2 | Gene names | Egr4 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 4 (EGR-4). | |||||
|
EGR1_HUMAN
|
||||||
| NC score | 0.003923 (rank : 107) | θ value | 6.88961 (rank : 92) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
SP5_MOUSE
|
||||||
| NC score | 0.003728 (rank : 108) | θ value | 2.36792 (rank : 64) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9JHX2 | Gene names | Sp5 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor Sp5. | |||||
|
SP5_HUMAN
|
||||||
| NC score | 0.003599 (rank : 109) | θ value | 3.0926 (rank : 72) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 832 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6BEB4 | Gene names | SP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor Sp5. | |||||
|
GLI1_HUMAN
|
||||||
| NC score | 0.003053 (rank : 110) | θ value | 6.88961 (rank : 93) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P08151, Q8TDN9 | Gene names | GLI1, GLI | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene) (Oncogene GLI). | |||||
|
ZBTB4_HUMAN
|
||||||
| NC score | 0.002799 (rank : 111) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 926 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P1Z0, Q7Z697, Q86XJ4, Q8N4V8 | Gene names | ZBTB4, KIAA1538 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 4 (KAISO-like zinc finger protein 1) (KAISO-L1). | |||||
|
CDON_MOUSE
|
||||||
| NC score | 0.002790 (rank : 112) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q32MD9, O88971 | Gene names | Cdon, Cdo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell adhesion molecule-related/down-regulated by oncogenes precursor. | |||||
|
DDX54_HUMAN
|
||||||
| NC score | 0.002192 (rank : 113) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 113 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TDD1, Q86YT8, Q9BRZ1 | Gene names | DDX54 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent RNA helicase DDX54 (EC 3.6.1.-) (DEAD box protein 54) (ATP-dependent RNA helicase DP97). | |||||