Please be patient as the page loads
|
RUNX3_HUMAN
|
||||||
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |
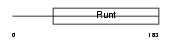
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RUNX3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.978505 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |
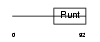
|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| θ value | 4.15819e-130 (rank : 3) | NC score | 0.957812 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_MOUSE
|
||||||
| θ value | 1.47893e-127 (rank : 4) | NC score | 0.960016 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |
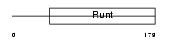
|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX2_HUMAN
|
||||||
| θ value | 1.09934e-114 (rank : 5) | NC score | 0.949771 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| θ value | 1.21547e-113 (rank : 6) | NC score | 0.945060 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
EP400_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.042405 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.048937 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.074374 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PHF1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.054459 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |
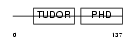
|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
ZIMP7_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.043980 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CNOT3_MOUSE
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.059089 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
HCN4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.032403 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.058814 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NRG3_MOUSE
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.054314 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35181 | Gene names | Nrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.062752 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.21417 (rank : 17) | NC score | 0.047213 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 18) | NC score | 0.062001 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
ZCH14_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.066419 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
NRG3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.055558 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56975 | Gene names | NRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.046241 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
RREB1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.010638 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
SNPH_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.032930 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15079 | Gene names | SNPH, KIAA0374 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaphilin. | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.018979 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
PDC6I_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.035119 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
PHF1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.047419 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
TBR1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 27) | NC score | 0.024555 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |
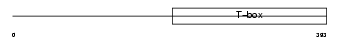
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
MAST1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.017428 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 29) | NC score | 0.017195 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
TBR1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 30) | NC score | 0.024185 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16650, Q53TH0 | Gene names | TBR1 | |||
|
Domain Architecture |
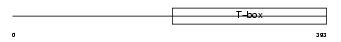
|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.012216 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.070291 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.071225 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.030846 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
EPN1_MOUSE
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.022485 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
MAST2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.014652 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.014434 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MTMR7_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.014021 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
SYNPO_MOUSE
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.040402 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
CN032_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.045705 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.006848 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
LRIG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.004078 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 627 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70193 | Gene names | Lrig1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.028515 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.016264 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
EGR1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.004958 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
GCM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.024940 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75603 | Gene names | GCM2, GCMB | |||
|
Domain Architecture |
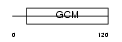
|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (hGCMb). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.035765 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.009623 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
RGPD8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 49) | NC score | 0.012542 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
SON_MOUSE
|
||||||
| θ value | 1.81305 (rank : 50) | NC score | 0.031066 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 1.81305 (rank : 51) | NC score | 0.044298 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
CT151_HUMAN
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.028859 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |
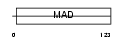
|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
EWS_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.036545 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
LZTS2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.013004 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
AB1IP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.022351 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.063261 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.033233 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
HSPB8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.016217 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |
|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
PITM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.010333 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
SMAD1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.009175 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.045937 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.045904 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.062795 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.063482 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.016449 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.026246 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.028776 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 3.0926 (rank : 68) | NC score | 0.021147 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TS101_MOUSE
|
||||||
| θ value | 3.0926 (rank : 69) | NC score | 0.029466 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
WASIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 70) | NC score | 0.029860 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
CI079_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.028024 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
CNOT3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.049638 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
HGS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.035389 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.035655 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.036496 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.032302 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
EWS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 77) | NC score | 0.021024 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
FGD1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 78) | NC score | 0.006698 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
NFRKB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 79) | NC score | 0.023859 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
AP2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.012790 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92481, Q5JYX6, Q9NQ63, Q9NU99, Q9UJI7, Q9Y214, Q9Y3K3 | Gene names | TFAP2B | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
AP2B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.012777 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61313 | Gene names | Tfap2b, Tcfap2b | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.038937 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.033362 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
NKTR_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.007328 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.021061 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PGCA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.008452 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
RNC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.019453 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
WBS14_MOUSE
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.014134 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
IDD_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.017903 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |
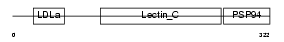
|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
MCR_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.002591 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||
|
NKX61_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.002745 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
PLXB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.004222 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
RTN4R_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.002757 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZR6 | Gene names | RTN4R, NOGOR | |||
|
Domain Architecture |
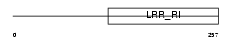
|
|||||
| Description | Reticulon-4 receptor precursor (Nogo receptor) (NgR) (Nogo-66 receptor). | |||||
|
TBX19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.013533 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99ME7 | Gene names | Tbx19, Tpit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.016517 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
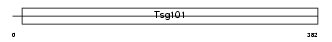
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
ZDHC8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.006349 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
ARI1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.068605 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.051137 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
RUNX3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q13761, Q12969, Q13760 | Gene names | RUNX3, AML2, CBFA3, PEBP2A3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX3_MOUSE
|
||||||
| NC score | 0.978505 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q64131, Q99P92, Q9R199 | Gene names | Runx3, Aml2, Cbfa3, Pebp2a3 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 3 (Core-binding factor, alpha 3 subunit) (CBF-alpha 3) (Acute myeloid leukemia 2 protein) (Oncogene AML-2) (Polyomavirus enhancer-binding protein 2 alpha C subunit) (PEBP2-alpha C) (PEA2-alpha C) (SL3-3 enhancer factor 1 alpha C subunit) (SL3/AKV core-binding factor alpha C subunit). | |||||
|
RUNX1_MOUSE
|
||||||
| NC score | 0.960016 (rank : 3) | θ value | 1.47893e-127 (rank : 4) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q03347, O08598, Q62049, Q9ESB9, Q9ET65 | Gene names | Runx1, Aml1, Cbfa2, Pebp2ab | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX1_HUMAN
|
||||||
| NC score | 0.957812 (rank : 4) | θ value | 4.15819e-130 (rank : 3) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q01196, O60472, O60473, O76047, O76089, Q13081, Q13755, Q13756, Q13757, Q13758, Q13759, Q15341, Q15343, Q16122, Q16284, Q16285, Q16286, Q16346, Q16347, Q92479 | Gene names | RUNX1, AML1, CBFA2 | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 1 (Core-binding factor, alpha 2 subunit) (CBF-alpha 2) (Acute myeloid leukemia 1 protein) (Oncogene AML-1) (Polyomavirus enhancer-binding protein 2 alpha B subunit) (PEBP2-alpha B) (PEA2-alpha B) (SL3-3 enhancer factor 1 alpha B subunit) (SL3/AKV core-binding factor alpha B subunit). | |||||
|
RUNX2_HUMAN
|
||||||
| NC score | 0.949771 (rank : 5) | θ value | 1.09934e-114 (rank : 5) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13950, O14614, O14615, O95181 | Gene names | RUNX2, AML3, CBFA1, OSF2, PEBP2A | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
RUNX2_MOUSE
|
||||||
| NC score | 0.945060 (rank : 6) | θ value | 1.21547e-113 (rank : 6) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q08775, O35183, Q08776, Q9JLN0, Q9QUQ6, Q9QY29, Q9R0U4, Q9Z2J7 | Gene names | Runx2, Aml3, Cbfa1, Osf2, Pebp2a | |||
|
Domain Architecture |

|
|||||
| Description | Runt-related transcription factor 2 (Core-binding factor, alpha 1 subunit) (CBF-alpha 1) (Acute myeloid leukemia 3 protein) (Oncogene AML-3) (Polyomavirus enhancer-binding protein 2 alpha A subunit) (PEBP2-alpha A) (PEA2-alpha A) (SL3-3 enhancer factor 1 alpha A subunit) (SL3/AKV core-binding factor alpha A subunit) (Osteoblast- specific transcription factor 2) (OSF-2). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.074374 (rank : 7) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.071225 (rank : 8) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.070291 (rank : 9) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
ARI1A_HUMAN
|
||||||
| NC score | 0.068605 (rank : 10) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 787 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O14497, Q53FK9, Q5T0W1, Q5T0W2, Q5T0W3, Q8NFD6, Q96T89, Q9BY33, Q9HBJ5, Q9UPZ1 | Gene names | ARID1A, C1orf4, OSA1, SMARCF1 | |||
|
Domain Architecture |

|
|||||
| Description | AT-rich interactive domain-containing protein 1A (ARID domain- containing protein 1A) (SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin subfamily F member 1) (SWI-SNF complex protein p270) (B120) (SWI-like protein) (Osa homolog 1) (hOSA1) (hELD) (BRG1-associated factor 250) (BAF250) (BRG1-associated factor 250a) (BAF250A). | |||||
|
ZCH14_MOUSE
|
||||||
| NC score | 0.066419 (rank : 11) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8VIG0, Q80TX3, Q91VU4 | Gene names | Zcchc14, Kiaa0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.063482 (rank : 12) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.063261 (rank : 13) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.062795 (rank : 14) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.062752 (rank : 15) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.062001 (rank : 16) | θ value | 0.21417 (rank : 18) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
CNOT3_MOUSE
|
||||||
| NC score | 0.059089 (rank : 17) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8K0V4 | Gene names | Cnot3, Not3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.058814 (rank : 18) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
NRG3_HUMAN
|
||||||
| NC score | 0.055558 (rank : 19) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56975 | Gene names | NRG3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
PHF1_MOUSE
|
||||||
| NC score | 0.054459 (rank : 20) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1B8, O54808 | Gene names | Phf1, Tctex-3, Tctex3 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1) (T-complex testis-expressed 3) (Polycomblike 1) (mPCl1). | |||||
|
NRG3_MOUSE
|
||||||
| NC score | 0.054314 (rank : 21) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35181 | Gene names | Nrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pro-neuregulin-3, membrane-bound isoform precursor (Pro-NRG3) [Contains: Neuregulin-3 (NRG-3)]. | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.051137 (rank : 22) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CNOT3_HUMAN
|
||||||
| NC score | 0.049638 (rank : 23) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75175, Q9NZN7, Q9UF76 | Gene names | CNOT3, KIAA0691, NOT3 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 3 (CCR4-associated factor 3). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.048937 (rank : 24) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
PHF1_HUMAN
|
||||||
| NC score | 0.047419 (rank : 25) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43189, O60929, Q96KM7 | Gene names | PHF1 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 1 (Protein PHF1). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.047213 (rank : 26) | θ value | 0.21417 (rank : 17) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.046241 (rank : 27) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.045937 (rank : 28) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.045904 (rank : 29) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.045705 (rank : 30) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.044298 (rank : 31) | θ value | 1.81305 (rank : 51) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZIMP7_HUMAN
|
||||||
| NC score | 0.043980 (rank : 32) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8NF64, O94790, Q659A8, Q6JKL5, Q8WTX8, Q96Q01, Q9BQH7 | Gene names | ZIMP7, KIAA1886 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
EP400_HUMAN
|
||||||
| NC score | 0.042405 (rank : 33) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q96L91, O15411, Q6P2F5, Q8N8Q7, Q8NE05, Q96JK7, Q9P230 | Gene names | EP400, CAGH32, KIAA1498, KIAA1818, TNRC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (hDomino) (CAG repeat protein 32) (Trinucleotide repeat-containing gene 12 protein). | |||||
|
SYNPO_MOUSE
|
||||||
| NC score | 0.040402 (rank : 34) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8CC35, Q99JI0 | Gene names | Synpo, Kiaa1029 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin. | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.038937 (rank : 35) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
EWS_MOUSE
|
||||||
| NC score | 0.036545 (rank : 36) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61545 | Gene names | Ewsr1, Ews, Ewsh | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS. | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.036496 (rank : 37) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.035765 (rank : 38) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.035655 (rank : 39) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
HGS_MOUSE
|
||||||
| NC score | 0.035389 (rank : 40) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 589 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q99LI8, Q61691, Q8BQW3 | Gene names | Hgs, Hrs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate. | |||||
|
PDC6I_HUMAN
|
||||||
| NC score | 0.035119 (rank : 41) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WUM4, Q9BX86, Q9NUN0, Q9P2H2, Q9UKL5 | Gene names | PDCD6IP, AIP1, ALIX, KIAA1375 | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death 6-interacting protein (PDCD6-interacting protein) (ALG-2-interacting protein 1) (Hp95). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.033362 (rank : 42) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.033233 (rank : 43) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
SNPH_HUMAN
|
||||||
| NC score | 0.032930 (rank : 44) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15079 | Gene names | SNPH, KIAA0374 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Syntaphilin. | |||||
|
HCN4_HUMAN
|
||||||
| NC score | 0.032403 (rank : 45) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9Y3Q4, Q9UMQ7 | Gene names | HCN4 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4. | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.032302 (rank : 46) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.031066 (rank : 47) | θ value | 1.81305 (rank : 50) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.030846 (rank : 48) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.029860 (rank : 49) | θ value | 3.0926 (rank : 70) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
TS101_MOUSE
|
||||||
| NC score | 0.029466 (rank : 50) | θ value | 3.0926 (rank : 69) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61187 | Gene names | Tsg101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
CT151_HUMAN
|
||||||
| NC score | 0.028859 (rank : 51) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NC74, Q8N4Z9, Q9BR75, Q9H0Y9 | Gene names | C20orf151 | |||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf151. | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.028776 (rank : 52) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.028515 (rank : 53) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
CI079_HUMAN
|
||||||
| NC score | 0.028024 (rank : 54) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6ZUB1, Q5SQC9, Q8NA41, Q8ND27 | Gene names | C9orf79 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf79. | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.026246 (rank : 55) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
GCM2_HUMAN
|
||||||
| NC score | 0.024940 (rank : 56) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O75603 | Gene names | GCM2, GCMB | |||
|
Domain Architecture |

|
|||||
| Description | Chorion-specific transcription factor GCMb (Glial cells missing homolog 2) (GCM motif protein 2) (hGCMb). | |||||
|
TBR1_MOUSE
|
||||||
| NC score | 0.024555 (rank : 57) | θ value | 0.47712 (rank : 27) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64336 | Gene names | Tbr1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
TBR1_HUMAN
|
||||||
| NC score | 0.024185 (rank : 58) | θ value | 0.62314 (rank : 30) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16650, Q53TH0 | Gene names | TBR1 | |||
|
Domain Architecture |

|
|||||
| Description | T-brain-1 protein (T-box brain protein 1) (TBR-1) (TES-56). | |||||
|
NFRKB_HUMAN
|
||||||
| NC score | 0.023859 (rank : 59) | θ value | 5.27518 (rank : 79) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6P4R8, Q12869, Q15312, Q9H048 | Gene names | NFRKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear factor related to kappa-B-binding protein (DNA-binding protein R kappa-B). | |||||
|
EPN1_MOUSE
|
||||||
| NC score | 0.022485 (rank : 60) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q80VP1, O70446 | Gene names | Epn1 | |||
|
Domain Architecture |

|
|||||
| Description | Epsin-1 (EPS-15-interacting protein 1) (Intersectin-EH-binding protein 1) (Ibp1). | |||||
|
AB1IP_HUMAN
|
||||||
| NC score | 0.022351 (rank : 61) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q7Z5R6, Q8IWS8, Q8IZZ7 | Gene names | APBB1IP, PREL1, RARP1, RIAM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 1- interacting protein (APBB1-interacting protein 1) (Rap1-GTP- interacting adapter molecule) (RIAM) (Proline-rich EVH1 ligand 1) (PREL-1) (Proline-rich protein 73) (Retinoic acid-responsive proline- rich protein 1) (RARP-1). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.021147 (rank : 62) | θ value | 3.0926 (rank : 68) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.021061 (rank : 63) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
EWS_HUMAN
|
||||||
| NC score | 0.021024 (rank : 64) | θ value | 5.27518 (rank : 77) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q01844, Q92635 | Gene names | EWSR1, EWS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein EWS (EWS oncogene) (Ewing sarcoma breakpoint region 1 protein). | |||||
|
RNC_HUMAN
|
||||||
| NC score | 0.019453 (rank : 65) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRR4, Q7Z5V2, Q86YH0, Q9NW73, Q9Y2V9, Q9Y4Y0 | Gene names | RNASEN, RN3, RNASE3L | |||
|
Domain Architecture |

|
|||||
| Description | Ribonuclease III (EC 3.1.26.3) (RNase III) (Drosha) (p241). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.018979 (rank : 66) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
IDD_HUMAN
|
||||||
| NC score | 0.017903 (rank : 67) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98153 | Gene names | DGCR2, IDD, KIAA0163 | |||
|
Domain Architecture |

|
|||||
| Description | Integral membrane protein DGCR2/IDD precursor. | |||||
|
MAST1_HUMAN
|
||||||
| NC score | 0.017428 (rank : 68) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1146 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9Y2H9, O00114, Q8N6X0 | Gene names | MAST1, KIAA0973 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
MAST1_MOUSE
|
||||||
| NC score | 0.017195 (rank : 69) | θ value | 0.62314 (rank : 29) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1123 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9R1L5, Q7TQG9, Q80TN0 | Gene names | Mast1, Kiaa0973, Sast | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 1 (EC 2.7.11.1) (Syntrophin-associated serine/threonine-protein kinase). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.016517 (rank : 70) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.016449 (rank : 71) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.016264 (rank : 72) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
HSPB8_MOUSE
|
||||||
| NC score | 0.016217 (rank : 73) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JK92 | Gene names | Hspb8, Cryac, Hsp22 | |||
|
Domain Architecture |

|
|||||
| Description | Heat-shock protein beta-8 (HspB8) (Alpha crystallin C chain) (Small stress protein-like protein HSP22). | |||||
|
MAST2_HUMAN
|
||||||
| NC score | 0.014652 (rank : 74) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1189 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P0Q8, O94899, Q5VT07, Q5VT08, Q7LGC4, Q8NDG1, Q96B94, Q9BYE8 | Gene names | MAST2, KIAA0807, MAST205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
MAST2_MOUSE
|
||||||
| NC score | 0.014434 (rank : 75) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1110 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60592, Q6P9M1, Q8CHD1 | Gene names | Mast2, Mast205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 2 (EC 2.7.11.1). | |||||
|
WBS14_MOUSE
|
||||||
| NC score | 0.014134 (rank : 76) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99MZ3, Q99MY9, Q99MZ0, Q99MZ1, Q99MZ2, Q9JLM5 | Gene names | Mlxipl, Mio, Wbscr14 | |||
|
Domain Architecture |

|
|||||
| Description | Williams-Beuren syndrome chromosome region 14 protein homolog (Mlx interactor) (MLX-interacting protein-like). | |||||
|
MTMR7_MOUSE
|
||||||
| NC score | 0.014021 (rank : 77) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2C9, Q8C4J6 | Gene names | Mtmr7 | |||
|
Domain Architecture |

|
|||||
| Description | Myotubularin-related protein 7 (EC 3.1.3.-). | |||||
|
TBX19_MOUSE
|
||||||
| NC score | 0.013533 (rank : 78) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99ME7 | Gene names | Tbx19, Tpit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX19 (T-box protein 19) (T-box factor, pituitary). | |||||
|
LZTS2_MOUSE
|
||||||
| NC score | 0.013004 (rank : 79) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91YU6, Q569X6 | Gene names | Lzts2, Kiaa1813 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2. | |||||
|
AP2B_HUMAN
|
||||||
| NC score | 0.012790 (rank : 80) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92481, Q5JYX6, Q9NQ63, Q9NU99, Q9UJI7, Q9Y214, Q9Y3K3 | Gene names | TFAP2B | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
AP2B_MOUSE
|
||||||
| NC score | 0.012777 (rank : 81) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61313 | Gene names | Tfap2b, Tcfap2b | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor AP-2 beta (AP2-beta) (Activating enhancer-binding protein 2 beta). | |||||
|
RGPD8_HUMAN
|
||||||
| NC score | 0.012542 (rank : 82) | θ value | 1.81305 (rank : 49) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 135 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99666 | Gene names | RGPD8, RANBP2L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RANBP2-like and GRIP domain-containing protein 8 (Ran-binding protein 2-like 1) (RanBP2L1) (Sperm membrane protein BS-63). | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.012216 (rank : 83) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
RREB1_HUMAN
|
||||||
| NC score | 0.010638 (rank : 84) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 879 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92766, O75567 | Gene names | RREB1 | |||
|
Domain Architecture |

|
|||||
| Description | RAS-responsive element-binding protein 1 (RREB-1) (Raf-responsive zinc finger protein LZ321). | |||||
|
PITM1_MOUSE
|
||||||
| NC score | 0.010333 (rank : 85) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O35954 | Gene names | Pitpnm1, Dres9, Mpt1, Nir2, Pitpnm | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated phosphatidylinositol transfer protein 1 (Phosphatidylinositol transfer protein, membrane-associated 1) (PITPnm 1) (Mpt-1) (Pyk2 N-terminal domain-interacting receptor 2) (NIR-2) (Drosophila retinal degeneration B homolog 1) (RdgB1). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.009623 (rank : 86) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
SMAD1_HUMAN
|
||||||
| NC score | 0.009175 (rank : 87) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15797, Q16636 | Gene names | SMAD1, BSP1, MADH1, MADR1 | |||
|
Domain Architecture |

|
|||||
| Description | Mothers against decapentaplegic homolog 1 (SMAD 1) (Mothers against DPP homolog 1) (Mad-related protein 1) (Transforming growth factor- beta signaling protein 1) (BSP-1) (hSMAD1) (JV4-1). | |||||
|
PGCA_MOUSE
|
||||||
| NC score | 0.008452 (rank : 88) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61282, Q64021 | Gene names | Agc1, Agc | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP). | |||||
|
NKTR_MOUSE
|
||||||
| NC score | 0.007328 (rank : 89) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P30415 | Gene names | Nktr | |||
|
Domain Architecture |

|
|||||
| Description | NK-tumor recognition protein (Natural-killer cells cyclophilin-related protein) (NK-TR protein). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.006848 (rank : 90) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
FGD1_HUMAN
|
||||||
| NC score | 0.006698 (rank : 91) | θ value | 5.27518 (rank : 78) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 367 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P98174, Q8N4D9 | Gene names | FGD1, ZFYVE3 | |||
|
Domain Architecture |

|
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 1 (Faciogenital dysplasia 1 protein) (Zinc finger FYVE domain-containing protein 3) (Rho/Rac guanine nucleotide exchange factor FGD1) (Rho/Rac GEF). | |||||
|
ZDHC8_MOUSE
|
||||||
| NC score | 0.006349 (rank : 92) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q5Y5T5, Q7TNF7 | Gene names | Zdhhc8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable palmitoyltransferase ZDHHC8 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 8) (DHHC-8). | |||||
|
EGR1_HUMAN
|
||||||
| NC score | 0.004958 (rank : 93) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P18146 | Gene names | EGR1, ZNF225 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A) (Transcription factor ETR103) (Zinc finger protein 225) (AT225). | |||||
|
PLXB1_MOUSE
|
||||||
| NC score | 0.004222 (rank : 94) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8CJH3, Q6ZQC3, Q80ZZ1 | Gene names | Plxnb1, Kiaa0407 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin-B1 precursor. | |||||
|
LRIG1_MOUSE
|
||||||
| NC score | 0.004078 (rank : 95) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 627 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70193 | Gene names | Lrig1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and immunoglobulin-like domains protein 1 precursor (LIG-1). | |||||
|
RTN4R_HUMAN
|
||||||
| NC score | 0.002757 (rank : 96) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZR6 | Gene names | RTN4R, NOGOR | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 receptor precursor (Nogo receptor) (NgR) (Nogo-66 receptor). | |||||
|
NKX61_HUMAN
|
||||||
| NC score | 0.002745 (rank : 97) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P78426 | Gene names | NKX6-1, NKX6A | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-6.1. | |||||
|
MCR_MOUSE
|
||||||
| NC score | 0.002591 (rank : 98) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 96 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8VII8, Q8VII9 | Gene names | Nr3c2, Mlr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mineralocorticoid receptor (MR). | |||||