Please be patient as the page loads
|
NFAC3_HUMAN
|
||||||
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NFAC3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989556 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_MOUSE
|
||||||
| θ value | 3.87392e-144 (rank : 3) | NC score | 0.950830 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 8.0776e-142 (rank : 4) | NC score | 0.917550 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | 2.35023e-141 (rank : 5) | NC score | 0.939824 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | 8.41738e-115 (rank : 6) | NC score | 0.927376 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 1.09934e-114 (rank : 7) | NC score | 0.931743 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 7.73779e-60 (rank : 8) | NC score | 0.831471 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 1.45929e-58 (rank : 9) | NC score | 0.813186 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
TF65_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 10) | NC score | 0.261339 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| θ value | 2.61198e-07 (rank : 11) | NC score | 0.250008 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 0.000461057 (rank : 12) | NC score | 0.110949 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 13) | NC score | 0.104678 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
K1683_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 14) | NC score | 0.076577 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 15) | NC score | 0.057228 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 16) | NC score | 0.062410 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
EYA4_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.040275 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EYA4_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 18) | NC score | 0.041184 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
EGLN_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.042575 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63961, Q61520 | Gene names | Eng, Edg | |||
|
Domain Architecture |

|
|||||
| Description | Endoglin precursor (Cell surface MJ7/18 antigen). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 0.21417 (rank : 20) | NC score | 0.032451 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
SENP1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.026569 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 22) | NC score | 0.022011 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.033627 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
DCX_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.035300 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
R3HD2_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.069328 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
TTLL5_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.036238 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
TDRD7_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.024004 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
TRIM8_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.021932 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
SAM10_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.061589 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYL1 | Gene names | SAMD10, C20orf136 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
SAM10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.060844 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TST3 | Gene names | Samd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
TAB2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.026296 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.06291 (rank : 32) | NC score | 0.038468 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
ZCH14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 33) | NC score | 0.025129 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 1.38821 (rank : 34) | NC score | 0.042169 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
IRS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 35) | NC score | 0.025734 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |
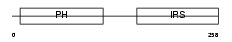
|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRS1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 36) | NC score | 0.025985 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 37) | NC score | 0.040749 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
WNK3_HUMAN
|
||||||
| θ value | 1.38821 (rank : 38) | NC score | 0.007052 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
CAC1F_HUMAN
|
||||||
| θ value | 1.81305 (rank : 39) | NC score | 0.008203 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
PHF12_HUMAN
|
||||||
| θ value | 1.81305 (rank : 40) | NC score | 0.019313 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
SPT5H_HUMAN
|
||||||
| θ value | 1.81305 (rank : 41) | NC score | 0.034725 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
SPT5H_MOUSE
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.034809 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.030660 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
DCX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.027898 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |
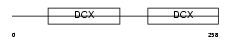
|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
MILK1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.015286 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MKL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.014525 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
WDR53_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.010675 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB94 | Gene names | Wdr53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 53. | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.001500 (rank : 106) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.049005 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DMRTD_HUMAN
|
||||||
| θ value | 3.0926 (rank : 50) | NC score | 0.021831 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
ELL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.014206 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
POF1B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.014489 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.018290 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TRIM8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.017739 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
APBB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.010453 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.033969 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
MAGI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.005828 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
PO6F2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.019064 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
SC24C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.024667 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
SMAP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.009799 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
AXUD1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.010250 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.004591 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
GON4L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.012899 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.013274 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
HNF1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.010112 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35680 | Gene names | TCF2, HNF1B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-beta (HNF-1beta) (HNF-1B) (Variant hepatic nuclear factor 1) (VHNF1) (Homeoprotein LFB3) (Transcription factor 2) (TCF-2). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.020301 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.019628 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PTC1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.013101 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
RIPK3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.003769 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |
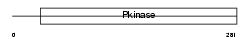
|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
SP100_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.013078 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35892, O35897, O88392, O88395 | Gene names | Sp100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein). | |||||
|
SPRM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.007221 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N7G0, Q8N748 | Gene names | SPRM1 | |||
|
Domain Architecture |
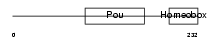
|
|||||
| Description | Sperm 1 POU-domain transcription factor (SPRM-1). | |||||
|
TENS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.019033 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
UBQL3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | 0.008958 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
CAC1B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.011000 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
CK024_HUMAN
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.031587 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
DCP1A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.012998 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
DYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.005760 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
FA61A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.016853 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
I18RA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.011291 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2B1 | Gene names | Il18rap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-18 receptor accessory protein precursor (IL-18 receptor accessory protein) (IL-18RAcP) (Interleukin-18 receptor accessory protein-like) (IL-18Rbeta) (IL-1R accessory protein-like) (IL-1RAcPL) (Accessory protein-like) (AcPL) (IL-1R7). | |||||
|
OBSCN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.002990 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
PHF12_MOUSE
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.015698 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
REPS2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.008108 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
SIM2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.005620 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
TAU_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.019066 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.017289 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ZN385_HUMAN
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.009361 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 8.99809 (rank : 87) | NC score | 0.003715 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
EAF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 88) | NC score | 0.009465 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
FA61A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.012927 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
INADL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.003454 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
JIP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.007562 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.008970 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
MRC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.002704 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.028204 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PODXL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.022178 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
RUSC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.009376 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
SPRE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.005851 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
SYP2L_MOUSE
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.010860 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
TRIP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.012673 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
TSSC4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.011278 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHE7 | Gene names | Tssc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4. | |||||
|
VGLL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.010962 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8G2 | Gene names | VGLL2, VITO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 2 (Vgl-2) (Protein VITO1). | |||||
|
ZN335_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | -0.001740 (rank : 107) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||
|
NBR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.051228 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15453 | Gene names | NBR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NBR2 (Next to BRCA1 gene 2 protein). | |||||
|
RELB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.098358 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
RELB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.094766 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |
|
|||||
| Description | Transcription factor RelB. | |||||
|
REL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.082227 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.070308 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.989556 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_MOUSE
|
||||||
| NC score | 0.950830 (rank : 3) | θ value | 3.87392e-144 (rank : 3) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.939824 (rank : 4) | θ value | 2.35023e-141 (rank : 5) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.931743 (rank : 5) | θ value | 1.09934e-114 (rank : 7) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.927376 (rank : 6) | θ value | 8.41738e-115 (rank : 6) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.917550 (rank : 7) | θ value | 8.0776e-142 (rank : 4) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.831471 (rank : 8) | θ value | 7.73779e-60 (rank : 8) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.813186 (rank : 9) | θ value | 1.45929e-58 (rank : 9) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.261339 (rank : 10) | θ value | 5.26297e-08 (rank : 10) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.250008 (rank : 11) | θ value | 2.61198e-07 (rank : 11) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.110949 (rank : 12) | θ value | 0.000461057 (rank : 12) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| NC score | 0.104678 (rank : 13) | θ value | 0.00228821 (rank : 13) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
RELB_HUMAN
|
||||||
| NC score | 0.098358 (rank : 14) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
RELB_MOUSE
|
||||||
| NC score | 0.094766 (rank : 15) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB. | |||||
|
REL_HUMAN
|
||||||
| NC score | 0.082227 (rank : 16) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.076577 (rank : 17) | θ value | 0.0148317 (rank : 14) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.070308 (rank : 18) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
R3HD2_MOUSE
|
||||||
| NC score | 0.069328 (rank : 19) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 397 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q80TM6, Q80YB1, Q8BLS5, Q9CW50 | Gene names | R3hdm2, Kiaa1002 | |||
|
Domain Architecture |

|
|||||
| Description | R3H domain-containing protein 2. | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.062410 (rank : 20) | θ value | 0.0431538 (rank : 16) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
SAM10_HUMAN
|
||||||
| NC score | 0.061589 (rank : 21) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYL1 | Gene names | SAMD10, C20orf136 | |||
|
Domain Architecture |

|
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
SAM10_MOUSE
|
||||||
| NC score | 0.060844 (rank : 22) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7TST3 | Gene names | Samd10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 10. | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.057228 (rank : 23) | θ value | 0.0148317 (rank : 15) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
NBR2_HUMAN
|
||||||
| NC score | 0.051228 (rank : 24) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15453 | Gene names | NBR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein NBR2 (Next to BRCA1 gene 2 protein). | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.049005 (rank : 25) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
EGLN_MOUSE
|
||||||
| NC score | 0.042575 (rank : 26) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q63961, Q61520 | Gene names | Eng, Edg | |||
|
Domain Architecture |

|
|||||
| Description | Endoglin precursor (Cell surface MJ7/18 antigen). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.042169 (rank : 27) | θ value | 1.38821 (rank : 34) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
EYA4_MOUSE
|
||||||
| NC score | 0.041184 (rank : 28) | θ value | 0.0563607 (rank : 18) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z191 | Gene names | Eya4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.040749 (rank : 29) | θ value | 1.38821 (rank : 37) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
EYA4_HUMAN
|
||||||
| NC score | 0.040275 (rank : 30) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O95677, O95464, O95679, Q8IW39, Q9NTR7 | Gene names | EYA4 | |||
|
Domain Architecture |

|
|||||
| Description | Eyes absent homolog 4 (EC 3.1.3.48). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.038468 (rank : 31) | θ value | 1.06291 (rank : 32) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
TTLL5_HUMAN
|
||||||
| NC score | 0.036238 (rank : 32) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6EMB2, Q9P1V5, Q9UPZ4 | Gene names | TTLL5, KIAA0998, STAMP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5 (SRC1 and TIF2-associated modulatory protein). | |||||
|
DCX_MOUSE
|
||||||
| NC score | 0.035300 (rank : 33) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88809 | Gene names | Dcx, Dcn | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
SPT5H_MOUSE
|
||||||
| NC score | 0.034809 (rank : 34) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 437 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O55201, Q3UJ77, Q3UJH1, Q3UKD7, Q3UM54, Q6PB73, Q6PDP0, Q6PFR4 | Gene names | Supt5h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (DRB sensitivity-inducing factor large subunit) (DSIF large subunit). | |||||
|
SPT5H_HUMAN
|
||||||
| NC score | 0.034725 (rank : 35) | θ value | 1.81305 (rank : 41) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O00267, O43279, Q59G52, Q99639 | Gene names | SUPT5H, SPT5, SPT5H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor SPT5 (hSPT5) (DRB sensitivity-inducing factor large subunit) (DSIF large subunit) (DSIF p160) (Tat- cotransactivator 1 protein) (Tat-CT1 protein). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.033969 (rank : 36) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.033627 (rank : 37) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.032451 (rank : 38) | θ value | 0.21417 (rank : 20) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CK024_HUMAN
|
||||||
| NC score | 0.031587 (rank : 39) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q96F05, Q9H2K4 | Gene names | C11orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C11orf24 precursor (Protein DM4E3). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.030660 (rank : 40) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.028204 (rank : 41) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
DCX_HUMAN
|
||||||
| NC score | 0.027898 (rank : 42) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O43602, O43911 | Gene names | DCX, DBCN, LISX | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal migration protein doublecortin (Lissencephalin-X) (Lis-X) (Doublin). | |||||
|
SENP1_HUMAN
|
||||||
| NC score | 0.026569 (rank : 43) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0U3, Q86XC8 | Gene names | SENP1 | |||
|
Domain Architecture |

|
|||||
| Description | Sentrin-specific protease 1 (EC 3.4.22.-) (Sentrin/SUMO-specific protease SENP1). | |||||
|
TAB2_MOUSE
|
||||||
| NC score | 0.026296 (rank : 44) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99K90, Q3UGP1, Q8BTP4, Q8CHD3, Q99KP4 | Gene names | Map3k7ip2, Kiaa0733, Tab2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2) (TAB2). | |||||
|
IRS1_MOUSE
|
||||||
| NC score | 0.025985 (rank : 45) | θ value | 1.38821 (rank : 36) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35569 | Gene names | Irs1, Irs-1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
IRS1_HUMAN
|
||||||
| NC score | 0.025734 (rank : 46) | θ value | 1.38821 (rank : 35) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35568 | Gene names | IRS1 | |||
|
Domain Architecture |

|
|||||
| Description | Insulin receptor substrate 1 (IRS-1). | |||||
|
ZCH14_HUMAN
|
||||||
| NC score | 0.025129 (rank : 47) | θ value | 1.06291 (rank : 33) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WYQ9, O60324, Q9UFP0 | Gene names | ZCCHC14, KIAA0579 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 14 (BDG-29). | |||||
|
SC24C_HUMAN
|
||||||
| NC score | 0.024667 (rank : 48) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P53992, Q8WV25 | Gene names | SEC24C, KIAA0079 | |||
|
Domain Architecture |

|
|||||
| Description | Protein transport protein Sec24C (SEC24-related protein C). | |||||
|
TDRD7_HUMAN
|
||||||
| NC score | 0.024004 (rank : 49) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NHU6, Q96JT1, Q9UFF0, Q9Y2M3 | Gene names | TDRD7, PCTAIRE2BP | |||
|
Domain Architecture |

|
|||||
| Description | Tudor domain-containing protein 7 (Tudor repeat associator with PCTAIRE 2) (Trap). | |||||
|
PODXL_HUMAN
|
||||||
| NC score | 0.022178 (rank : 50) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O00592 | Gene names | PODXL, PCLP, PCLP1 | |||
|
Domain Architecture |

|
|||||
| Description | Podocalyxin-like protein 1 precursor. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.022011 (rank : 51) | θ value | 0.279714 (rank : 22) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
TRIM8_MOUSE
|
||||||
| NC score | 0.021932 (rank : 52) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99PJ2, Q8C508, Q8C700, Q8CGI2, Q99PQ4 | Gene names | Trim8, Gerp, Rnf27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
DMRTD_HUMAN
|
||||||
| NC score | 0.021831 (rank : 53) | θ value | 3.0926 (rank : 50) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IXT2, Q8N6Q2, Q96M39, Q96SD4 | Gene names | DMRTC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Doublesex- and mab-3-related transcription factor C2. | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.020301 (rank : 54) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.019628 (rank : 55) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PHF12_HUMAN
|
||||||
| NC score | 0.019313 (rank : 56) | θ value | 1.81305 (rank : 40) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96QT6, Q6ZML2, Q9BV34, Q9H7U9, Q9P205 | Gene names | PHF12, KIAA1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.019066 (rank : 57) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
PO6F2_HUMAN
|
||||||
| NC score | 0.019064 (rank : 58) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P78424, P78425, Q75ME8, Q86UM6, Q9UDS7 | Gene names | POU6F2, RPF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 6, transcription factor 2 (Retina-derived POU-domain factor 1) (RPF-1). | |||||
|
TENS4_MOUSE
|
||||||
| NC score | 0.019033 (rank : 59) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BZ33, Q3TCM8, Q7TNR5 | Gene names | Tns4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tensin-4 precursor. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.018290 (rank : 60) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TRIM8_HUMAN
|
||||||
| NC score | 0.017739 (rank : 61) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BZR9, Q9C028 | Gene names | TRIM8, GERP, RNF27 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 8 (RING finger protein 27) (Glioblastoma-expressed RING finger protein). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.017289 (rank : 62) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
FA61A_HUMAN
|
||||||
| NC score | 0.016853 (rank : 63) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8ND56, Q76LX7, Q96AR3, Q96K73, Q96SN5, Q9UFR3 | Gene names | FAM61A, C19orf13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A (Putative alpha synuclein-binding protein) (AlphaSNBP). | |||||
|
PHF12_MOUSE
|
||||||
| NC score | 0.015698 (rank : 64) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q5SPL2, Q5SPL3, Q6ZPN8, Q80W50 | Gene names | Phf12, Kiaa1523 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 12 (PHD factor 1) (Pf1). | |||||
|
MILK1_MOUSE
|
||||||
| NC score | 0.015286 (rank : 65) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 788 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BGT6, Q8BJ60 | Gene names | Mirab13, D15Mit260, Kiaa1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MKL1_HUMAN
|
||||||
| NC score | 0.014525 (rank : 66) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q969V6, Q8TCL1, Q96SC5, Q96SC6, Q9P2B0 | Gene names | MKL1, KIAA1438, MAL | |||
|
Domain Architecture |

|
|||||
| Description | MKL/myocardin-like protein 1 (Myocardin-related transcription factor A) (MRTF-A) (Megakaryoblastic leukemia 1 protein) (Megakaryocytic acute leukemia protein). | |||||
|
POF1B_HUMAN
|
||||||
| NC score | 0.014489 (rank : 67) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 494 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8WVV4, Q5H9E9, Q5H9F0, Q8NG12, Q9H5Y2, Q9H738, Q9H744 | Gene names | POF1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein POF1B (Premature ovarian failure protein 1B). | |||||
|
ELL_MOUSE
|
||||||
| NC score | 0.014206 (rank : 68) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08856 | Gene names | Ell | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II elongation factor ELL (Eleven-nineteen lysine-rich leukemia protein). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.013274 (rank : 69) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
PTC1_MOUSE
|
||||||
| NC score | 0.013101 (rank : 70) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q61115 | Gene names | Ptch1, Ptch | |||
|
Domain Architecture |

|
|||||
| Description | Protein patched homolog 1 (PTC1) (PTC). | |||||
|
SP100_MOUSE
|
||||||
| NC score | 0.013078 (rank : 71) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35892, O35897, O88392, O88395 | Gene names | Sp100 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigen Sp-100 (Speckled 100 kDa) (Nuclear dot-associated Sp100 protein). | |||||
|
DCP1A_MOUSE
|
||||||
| NC score | 0.012998 (rank : 72) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q91YD3, Q6NZE3 | Gene names | Dcp1a, Mitc1, Smif | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | mRNA decapping enzyme 1A (EC 3.-.-.-) (Transcription factor SMIF) (MAD homolog 4-interacting transcription coactivator 1) (Smad4-interacting transcriptional co-activator). | |||||
|
FA61A_MOUSE
|
||||||
| NC score | 0.012927 (rank : 73) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K2F8, Q9CTG8 | Gene names | Fam61a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM61A. | |||||
|
GON4L_HUMAN
|
||||||
| NC score | 0.012899 (rank : 74) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 555 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q3T8J9, Q3T8J8, Q5VYZ5, Q5W0D5, Q6AWA6, Q6P1Q6, Q7Z3L3, Q8IY79, Q9BQI1, Q9HCG6 | Gene names | GON4L, GON4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
TRIP6_HUMAN
|
||||||
| NC score | 0.012673 (rank : 75) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15654, O15170, O15275, Q9BTB2, Q9UNT4 | Gene names | TRIP6, OIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 6 (TRIP6) (OPA-interacting protein 1) (Zyxin-related protein 1) (ZRP-1). | |||||
|
I18RA_MOUSE
|
||||||
| NC score | 0.011291 (rank : 76) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2B1 | Gene names | Il18rap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Interleukin-18 receptor accessory protein precursor (IL-18 receptor accessory protein) (IL-18RAcP) (Interleukin-18 receptor accessory protein-like) (IL-18Rbeta) (IL-1R accessory protein-like) (IL-1RAcPL) (Accessory protein-like) (AcPL) (IL-1R7). | |||||
|
TSSC4_MOUSE
|
||||||
| NC score | 0.011278 (rank : 77) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHE7 | Gene names | Tssc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC4. | |||||
|
CAC1B_MOUSE
|
||||||
| NC score | 0.011000 (rank : 78) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O55017, Q60609 | Gene names | Cacna1b, Cach5, Cacnl1a5, Cchn1a | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent N-type calcium channel subunit alpha-1B (Voltage- gated calcium channel subunit alpha Cav2.2) (Calcium channel, L type, alpha-1 polypeptide isoform 5) (Brain calcium channel III) (BIII). | |||||
|
VGLL2_HUMAN
|
||||||
| NC score | 0.010962 (rank : 79) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8G2 | Gene names | VGLL2, VITO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription cofactor vestigial-like protein 2 (Vgl-2) (Protein VITO1). | |||||
|
SYP2L_MOUSE
|
||||||
| NC score | 0.010860 (rank : 80) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8BWB1 | Gene names | Synpo2l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Synaptopodin 2-like protein. | |||||
|
WDR53_MOUSE
|
||||||
| NC score | 0.010675 (rank : 81) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DB94 | Gene names | Wdr53 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat protein 53. | |||||
|
APBB2_HUMAN
|
||||||
| NC score | 0.010453 (rank : 82) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92870, Q8IUI6 | Gene names | APBB2, FE65L, FE65L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 precursor protein-binding family B member 2 (Fe65-like protein). | |||||
|
AXUD1_MOUSE
|
||||||
| NC score | 0.010250 (rank : 83) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P59054 | Gene names | Axud1, Taip3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Axin-1 up-regulated gene 1 protein (TGF-beta-induced apoptosis protein 3) (TAIP-3). | |||||
|
HNF1B_HUMAN
|
||||||
| NC score | 0.010112 (rank : 84) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35680 | Gene names | TCF2, HNF1B | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 1-beta (HNF-1beta) (HNF-1B) (Variant hepatic nuclear factor 1) (VHNF1) (Homeoprotein LFB3) (Transcription factor 2) (TCF-2). | |||||
|
SMAP1_MOUSE
|
||||||
| NC score | 0.009799 (rank : 85) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q91VZ6, Q497I6, Q68EF3 | Gene names | Smap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stromal membrane-associated protein 1. | |||||
|
EAF1_HUMAN
|
||||||
| NC score | 0.009465 (rank : 86) | θ value | 8.99809 (rank : 88) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96JC9, Q8IW10 | Gene names | EAF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELL-associated factor 1. | |||||
|
RUSC2_MOUSE
|
||||||
| NC score | 0.009376 (rank : 87) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q80U22, Q8R1D6 | Gene names | Rusc2, Kiaa0375 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and SH3 domain-containing protein 2. | |||||
|
ZN385_HUMAN
|
||||||
| NC score | 0.009361 (rank : 88) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96PM9, Q5VH53, Q9H7R6, Q9UFU3 | Gene names | ZNF385, HZF, RZF | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 385 (Hematopoietic zinc finger protein) (Retinal zinc finger protein). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.008970 (rank : 89) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
UBQL3_MOUSE
|
||||||
| NC score | 0.008958 (rank : 90) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 120 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C5U9 | Gene names | Ubqln3 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-3. | |||||
|
CAC1F_HUMAN
|
||||||
| NC score | 0.008203 (rank : 91) | θ value | 1.81305 (rank : 39) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60840, O43901 | Gene names | CACNA1F, CACNAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1F (Voltage- gated calcium channel subunit alpha Cav1.4). | |||||
|
REPS2_HUMAN
|
||||||
| NC score | 0.008108 (rank : 92) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NFH8, O43428, Q8NFI5 | Gene names | REPS2, POB1 | |||
|
Domain Architecture |

|
|||||
| Description | RalBP1-associated Eps domain-containing protein 2 (RalBP1-interacting protein 2) (Partner of RalBP1). | |||||
|
JIP3_HUMAN
|
||||||
| NC score | 0.007562 (rank : 93) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1039 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9UPT6, Q96RY4, Q9H4I4, Q9H7P1, Q9NUG0 | Gene names | MAPK8IP3, JIP3, KIAA1066 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 3 (JNK-interacting protein 3) (JIP-3) (JNK MAP kinase scaffold protein 3) (Mitogen- activated protein kinase 8-interacting protein 3). | |||||
|
SPRM1_HUMAN
|
||||||
| NC score | 0.007221 (rank : 94) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N7G0, Q8N748 | Gene names | SPRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Sperm 1 POU-domain transcription factor (SPRM-1). | |||||
|
WNK3_HUMAN
|
||||||
| NC score | 0.007052 (rank : 95) | θ value | 1.38821 (rank : 38) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9BYP7, Q9HCK6 | Gene names | WNK3, KIAA1566, PRKWNK3 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK3 (EC 2.7.11.1) (Protein kinase with no lysine 3) (Protein kinase, lysine-deficient 3). | |||||
|
SPRE1_HUMAN
|
||||||
| NC score | 0.005851 (rank : 96) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z699, Q8N256 | Gene names | SPRED1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sprouty-related, EVH1 domain-containing protein 1 (Spred-1). | |||||
|
MAGI1_HUMAN
|
||||||
| NC score | 0.005828 (rank : 97) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96QZ7, O00309, O43863, O75085, Q96QZ8, Q96QZ9 | Gene names | MAGI1, BAIAP1, BAP1, TNRC19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1) (Atrophin-1-interacting protein 3) (AIP3) (WW domain-containing protein 3) (WWP3) (Trinucleotide repeat-containing gene 19 protein). | |||||
|
DYH11_HUMAN
|
||||||
| NC score | 0.005760 (rank : 98) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96DT5, Q9UJ82 | Gene names | DNAH11 | |||
|
Domain Architecture |

|
|||||
| Description | Ciliary dynein heavy chain 11 (Axonemal beta dynein heavy chain 11). | |||||
|
SIM2_MOUSE
|
||||||
| NC score | 0.005620 (rank : 99) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61079, O35391, Q61046, Q61904 | Gene names | Sim2 | |||
|
Domain Architecture |

|
|||||
| Description | Single-minded homolog 2 (SIM transcription factor) (mSIM). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.004591 (rank : 100) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
RIPK3_MOUSE
|
||||||
| NC score | 0.003769 (rank : 101) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 867 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QZL0 | Gene names | Ripk3, Rip3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-interacting serine/threonine-protein kinase 3 (EC 2.7.11.1) (RIP-like protein kinase 3) (Receptor-interacting protein 3) (RIP-3) (mRIP3). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.003715 (rank : 102) | θ value | 8.99809 (rank : 87) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
INADL_HUMAN
|
||||||
| NC score | 0.003454 (rank : 103) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NI35, O15249, O43742, O60833, Q5VUA5, Q5VUA6, Q5VUA7, Q5VUA8, Q5VUA9, Q5VUB0, Q8WU78, Q9H3N9 | Gene names | INADL, PATJ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | InaD-like protein (Inadl protein) (hINADL) (Pals1-associated tight junction protein) (Protein associated to tight junctions). | |||||
|
OBSCN_HUMAN
|
||||||
| NC score | 0.002990 (rank : 104) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5VST9, Q2A664, Q5T7G8, Q5T7G9, Q5VSU2, Q86YC7, Q8NHN0, Q8NHN1, Q8NHN2, Q8NHN4, Q8NHN5, Q8NHN6, Q8NHN7, Q8NHN8, Q8NHN9, Q96AA2, Q9HCD3, Q9HCL6 | Gene names | OBSCN, KIAA1556, KIAA1639 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Obscurin (Obscurin-myosin light chain kinase) (Obscurin-MLCK) (Obscurin-RhoGEF). | |||||
|
MRC1_HUMAN
|
||||||
| NC score | 0.002704 (rank : 105) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22897 | Gene names | MRC1 | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage mannose receptor 1 precursor (MMR) (CD206 antigen). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.001500 (rank : 106) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
ZN335_HUMAN
|
||||||
| NC score | -0.001740 (rank : 107) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 102 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H4Z2, Q9H684 | Gene names | ZNF335 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 335. | |||||