Please be patient as the page loads
|
REL_MOUSE
|
||||||
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
REL_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.977194 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
TF65_HUMAN
|
||||||
| θ value | 8.17181e-102 (rank : 3) | NC score | 0.932120 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| θ value | 4.05562e-101 (rank : 4) | NC score | 0.939917 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
RELB_MOUSE
|
||||||
| θ value | 4.08394e-77 (rank : 5) | NC score | 0.942292 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |
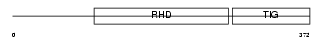
|
|||||
| Description | Transcription factor RelB. | |||||
|
RELB_HUMAN
|
||||||
| θ value | 1.5519e-76 (rank : 6) | NC score | 0.939747 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |
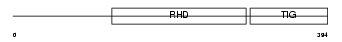
|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
NFKB2_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 7) | NC score | 0.553185 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_MOUSE
|
||||||
| θ value | 8.53533e-67 (rank : 8) | NC score | 0.551001 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25799, Q3TZE8, Q3V2V6, Q6TDG8, Q75ZL1, Q80Y21, Q8C712 | Gene names | Nfkb1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) (NF-kappa-B1 p84/NF-kappa-B1 p98) [Contains: Nuclear factor NF- kappa-B p50 subunit]. | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 1.11475e-66 (rank : 9) | NC score | 0.570363 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_HUMAN
|
||||||
| θ value | 1.90147e-66 (rank : 10) | NC score | 0.551075 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
CD45_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 11) | NC score | 0.023545 (rank : 125) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
ZIM10_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 12) | NC score | 0.050780 (rank : 103) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
SOX7_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.022422 (rank : 128) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 14) | NC score | 0.007836 (rank : 166) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
SYNJ1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 15) | NC score | 0.029627 (rank : 119) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 0.163984 (rank : 16) | NC score | 0.051162 (rank : 101) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
IRS2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 17) | NC score | 0.031842 (rank : 116) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
MANS1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 18) | NC score | 0.045773 (rank : 109) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8J5, Q8NEC1 | Gene names | MANSC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MANSC domain-containing protein 1 precursor. | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 0.21417 (rank : 19) | NC score | 0.044984 (rank : 110) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
NFIB_MOUSE
|
||||||
| θ value | 0.365318 (rank : 20) | NC score | 0.028119 (rank : 121) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97863, P70252, P70253, P70254, Q9R1G4 | Gene names | Nfib | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
CAC1C_HUMAN
|
||||||
| θ value | 0.47712 (rank : 21) | NC score | 0.010631 (rank : 154) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
PRG4_MOUSE
|
||||||
| θ value | 0.47712 (rank : 22) | NC score | 0.019961 (rank : 134) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 23) | NC score | 0.035081 (rank : 115) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 24) | NC score | 0.035114 (rank : 114) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 25) | NC score | 0.023492 (rank : 126) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ZIM10_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.043449 (rank : 111) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
NFIB_HUMAN
|
||||||
| θ value | 0.813845 (rank : 27) | NC score | 0.027381 (rank : 123) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |
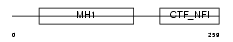
|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
BAT2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.035994 (rank : 112) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
CA051_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.029456 (rank : 120) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3TQ03, Q3U1B8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf51 homolog. | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 1.06291 (rank : 30) | NC score | 0.017788 (rank : 140) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 31) | NC score | 0.053149 (rank : 85) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
PCGF2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.023291 (rank : 127) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |
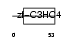
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
SIN3A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.029683 (rank : 118) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.020325 (rank : 133) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CLOCK_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.021055 (rank : 131) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
TTLL5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.016439 (rank : 144) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 37) | NC score | 0.027889 (rank : 122) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
AKAP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.015333 (rank : 146) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
GR65_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.017026 (rank : 141) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
PCGF2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.021610 (rank : 130) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |
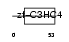
|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
PER3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.019409 (rank : 135) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
SIN3A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.030014 (rank : 117) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
TEX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 43) | NC score | 0.018775 (rank : 137) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 2.36792 (rank : 44) | NC score | 0.009228 (rank : 161) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.013218 (rank : 149) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
FOXI1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.008030 (rank : 165) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |
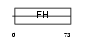
|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
TM108_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.020861 (rank : 132) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
AT10B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.005797 (rank : 168) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
FOXI1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.007821 (rank : 167) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q922I5, Q5SRI5, Q9D299 | Gene names | Foxi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I1. | |||||
|
GGA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.009625 (rank : 159) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
GP112_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.010534 (rank : 155) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
IF4G3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.009615 (rank : 160) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
LAP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.003289 (rank : 170) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
MINT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.011616 (rank : 152) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
NID1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.003678 (rank : 169) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
OLIG2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.012511 (rank : 151) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
AOC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.008449 (rank : 163) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75106, O00120, O75105, Q4TTW5, Q9UNY0 | Gene names | AOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retina-specific copper amine oxidase precursor (EC 1.4.3.6) (RAO) (Amine oxidase [copper-containing]). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.018719 (rank : 138) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
K0310_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.016990 (rank : 142) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MAML2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.015465 (rank : 145) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
MAVS_HUMAN
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.021926 (rank : 129) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
PRR8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.018894 (rank : 136) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
PTPRN_HUMAN
|
||||||
| θ value | 5.27518 (rank : 63) | NC score | 0.009194 (rank : 162) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
UBQL2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.008114 (rank : 164) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHD9, O94798, Q9H3W6, Q9HAZ4 | Gene names | UBQLN2, PLIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (hPLIC- 2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
ZIMP7_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.035227 (rank : 113) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
ATX7_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.013683 (rank : 148) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
ODO2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.011423 (rank : 153) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
SYNJ1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.023801 (rank : 124) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
ULK2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | -0.000433 (rank : 172) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||
|
VP13A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.013124 (rank : 150) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RL7, Q5JSX9, Q5JSY0, Q5VYR5, Q702P4, Q709D0, Q86YF8, Q96S61, Q9H995, Q9Y2J1 | Gene names | VPS13A, CHAC, KIAA0986 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 13A (Chorein) (Chorea- acanthocytosis protein). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.018323 (rank : 139) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CNOT4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.013756 (rank : 147) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
DPPA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.010238 (rank : 156) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWH0 | Gene names | Dppa2, Phsecrg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmental pluripotency-associated protein 2 (Pluripotent embryonic stem cell-related protein 1). | |||||
|
OLIG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.009962 (rank : 157) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
PGFRL_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.016552 (rank : 143) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PE55, Q3V182, Q9D0Z1 | Gene names | Pdgfrl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor receptor-like protein precursor. | |||||
|
PRDM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.000668 (rank : 171) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
WBP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.009772 (rank : 158) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97764, Q8WUP5, Q99J20 | Gene names | Wbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 1 (WBP-1). | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.052585 (rank : 88) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.057339 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.055669 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.056079 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055375 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.057984 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.057703 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.055655 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.056378 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANKR2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.051599 (rank : 97) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |
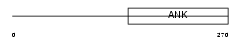
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
ANKR2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.050055 (rank : 107) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |
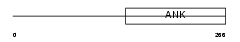
|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.067190 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANKR7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.059876 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |
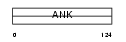
|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.060977 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
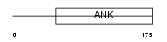
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.058880 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |
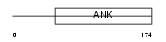
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.051498 (rank : 98) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
ANR23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.060373 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANR23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.056811 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANR25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.050047 (rank : 108) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.052041 (rank : 92) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ANR28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.054879 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.055118 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR29_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.050116 (rank : 106) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N6D5, Q6ZWE8, Q96LU9 | Gene names | ANKRD29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 29. | |||||
|
ANR33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.058307 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANR41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.056023 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
ANR42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.069573 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.056076 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.054631 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.052386 (rank : 90) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.058567 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.059752 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.057922 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
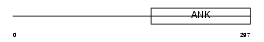
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.056981 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
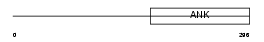
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.053589 (rank : 84) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
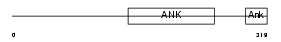
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.059154 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.051760 (rank : 95) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXK3, Q96EP7, Q9H8Z1 | Gene names | ASB13 | |||
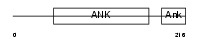
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 13 (ASB-13). | |||||
|
ASB13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.050362 (rank : 105) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBX0 | Gene names | Asb13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 13 (ASB-13). | |||||
|
ASB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.052486 (rank : 89) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
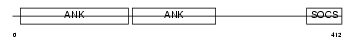
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
ASB15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.052096 (rank : 91) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ASB16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.058237 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |
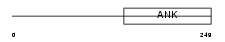
|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.068998 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.061155 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.062053 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.054379 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWX0 | Gene names | ASB5 | |||
|
Domain Architecture |
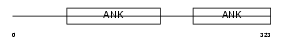
|
|||||
| Description | Ankyrin repeat and SOCS box protein 5 (ASB-5). | |||||
|
ASB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.064169 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |
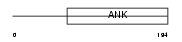
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.072142 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |
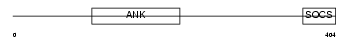
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.058434 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
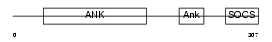
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.058325 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
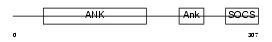
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.056798 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
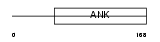
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.056285 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
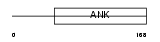
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.056842 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DX5, Q9NWS5, Q9Y4T3 | Gene names | ASB9 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
ASB9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.051434 (rank : 99) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT8, Q9DAF7 | Gene names | Asb9 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
BCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.116546 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |
|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.118123 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDN2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.051765 (rank : 94) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CDN2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.053942 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |
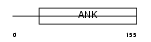
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.054536 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.056103 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.052607 (rank : 87) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAM9, Q9CUA7 | Gene names | Fank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains 1 protein (Germ cell- specific gene 1 protein) (GSG1). | |||||
|
GABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.051274 (rank : 100) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |
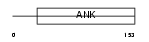
|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.051681 (rank : 96) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
IKBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.130830 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |
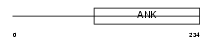
|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.131634 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.132251 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
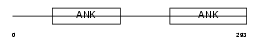
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.130086 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |
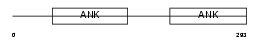
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
IKBE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.140875 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.134449 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
IKBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.075477 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |
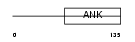
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
IKBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.073572 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |
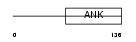
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
INVS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.053993 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
INVS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.052879 (rank : 86) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
MIB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.068587 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.068644 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.062049 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.058684 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.050883 (rank : 102) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.070308 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.072462 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.135622 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.141097 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
OSBL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.052000 (rank : 93) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
PA2G6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.070254 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PA2G6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.065808 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.068210 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
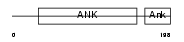
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.068100 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |
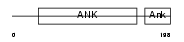
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
RFXK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.050528 (rank : 104) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14593, O95839 | Gene names | RFXANK, ANKRA1, RFXB | |||
|
Domain Architecture |
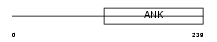
|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Ankyrin repeat family A protein 1). | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.068250 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.058995 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.060482 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.054040 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.055132 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ZDH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.057011 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |
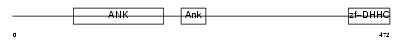
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZDH13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.056542 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |
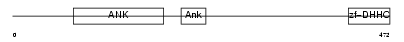
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.059889 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.058851 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |
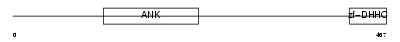
|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 77 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_HUMAN
|
||||||
| NC score | 0.977194 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
RELB_MOUSE
|
||||||
| NC score | 0.942292 (rank : 3) | θ value | 4.08394e-77 (rank : 5) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB. | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.939917 (rank : 4) | θ value | 4.05562e-101 (rank : 4) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
RELB_HUMAN
|
||||||
| NC score | 0.939747 (rank : 5) | θ value | 1.5519e-76 (rank : 6) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.932120 (rank : 6) | θ value | 8.17181e-102 (rank : 3) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.570363 (rank : 7) | θ value | 1.11475e-66 (rank : 9) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| NC score | 0.553185 (rank : 8) | θ value | 1.00825e-67 (rank : 7) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_HUMAN
|
||||||
| NC score | 0.551075 (rank : 9) | θ value | 1.90147e-66 (rank : 10) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
NFKB1_MOUSE
|
||||||
| NC score | 0.551001 (rank : 10) | θ value | 8.53533e-67 (rank : 8) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P25799, Q3TZE8, Q3V2V6, Q6TDG8, Q75ZL1, Q80Y21, Q8C712 | Gene names | Nfkb1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) (NF-kappa-B1 p84/NF-kappa-B1 p98) [Contains: Nuclear factor NF- kappa-B p50 subunit]. | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.141097 (rank : 11) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
IKBE_HUMAN
|
||||||
| NC score | 0.140875 (rank : 12) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.135622 (rank : 13) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.134449 (rank : 14) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.132251 (rank : 15) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBA_MOUSE
|
||||||
| NC score | 0.131634 (rank : 16) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBA_HUMAN
|
||||||
| NC score | 0.130830 (rank : 17) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBB_MOUSE
|
||||||
| NC score | 0.130086 (rank : 18) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.118123 (rank : 19) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
BCL3_HUMAN
|
||||||
| NC score | 0.116546 (rank : 20) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
IKBL_HUMAN
|
||||||
| NC score | 0.075477 (rank : 21) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
IKBL_MOUSE
|
||||||
| NC score | 0.073572 (rank : 22) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.072462 (rank : 23) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
ASB6_MOUSE
|
||||||
| NC score | 0.072142 (rank : 24) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.070308 (rank : 25) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
PA2G6_HUMAN
|
||||||
| NC score | 0.070254 (rank : 26) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ANR42_HUMAN
|
||||||
| NC score | 0.069573 (rank : 27) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ASB16_MOUSE
|
||||||
| NC score | 0.068998 (rank : 28) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.068644 (rank : 29) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.068587 (rank : 30) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.068250 (rank : 31) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.068210 (rank : 32) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| NC score | 0.068100 (rank : 33) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.067190 (rank : 34) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
PA2G6_MOUSE
|
||||||
| NC score | 0.065808 (rank : 35) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ASB6_HUMAN
|
||||||
| NC score | 0.064169 (rank : 36) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.062053 (rank : 37) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.062049 (rank : 38) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
ASB3_HUMAN
|
||||||
| NC score | 0.061155 (rank : 39) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ANR10_HUMAN
|
||||||
| NC score | 0.060977 (rank : 40) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.060482 (rank : 41) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
ANR23_HUMAN
|
||||||
| NC score | 0.060373 (rank : 42) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ZDH17_HUMAN
|
||||||
| NC score | 0.059889 (rank : 43) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ANKR7_HUMAN
|
||||||
| NC score | 0.059876 (rank : 44) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.059752 (rank : 45) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ASB10_MOUSE
|
||||||
| NC score | 0.059154 (rank : 46) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.058995 (rank : 47) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ANR10_MOUSE
|
||||||
| NC score | 0.058880 (rank : 48) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ZDH17_MOUSE
|
||||||
| NC score | 0.058851 (rank : 49) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.058684 (rank : 50) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.058567 (rank : 51) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ASB7_HUMAN
|
||||||
| NC score | 0.058434 (rank : 52) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| NC score | 0.058325 (rank : 53) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ANR33_HUMAN
|
||||||
| NC score | 0.058307 (rank : 54) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ASB16_HUMAN
|
||||||
| NC score | 0.058237 (rank : 55) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.057984 (rank : 56) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.057922 (rank : 57) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.057703 (rank : 58) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.057339 (rank : 59) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ZDH13_HUMAN
|
||||||
| NC score | 0.057011 (rank : 60) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.056981 (rank : 61) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB9_HUMAN
|
||||||
| NC score | 0.056842 (rank : 62) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DX5, Q9NWS5, Q9Y4T3 | Gene names | ASB9 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
ANR23_MOUSE
|
||||||
| NC score | 0.056811 (rank : 63) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ASB8_HUMAN
|
||||||
| NC score | 0.056798 (rank : 64) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ZDH13_MOUSE
|
||||||
| NC score | 0.056542 (rank : 65) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.056378 (rank : 66) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ASB8_MOUSE
|
||||||
| NC score | 0.056285 (rank : 67) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.056103 (rank : 68) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.056079 (rank : 69) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.056076 (rank : 70) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.056023 (rank : 71) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.055669 (rank : 72) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.055655 (rank : 73) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.055375 (rank : 74) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.055132 (rank : 75) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.055118 (rank : 76) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.054879 (rank : 77) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.054631 (rank : 78) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.054536 (rank : 79) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ASB5_HUMAN
|
||||||
| NC score | 0.054379 (rank : 80) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWX0 | Gene names | ASB5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 5 (ASB-5). | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.054040 (rank : 81) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.053993 (rank : 82) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
CDN2D_MOUSE
|
||||||
| NC score | 0.053942 (rank : 83) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
ASB10_HUMAN
|
||||||
| NC score | 0.053589 (rank : 84) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.053149 (rank : 85) | θ value | 1.06291 (rank : 31) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
INVS_MOUSE
|
||||||
| NC score | 0.052879 (rank : 86) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 609 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O89019, O88849 | Gene names | Invs, Inv, Nphp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning protein) (Nephrocystin-2). | |||||
|
FANK1_MOUSE
|
||||||
| NC score | 0.052607 (rank : 87) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAM9, Q9CUA7 | Gene names | Fank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains 1 protein (Germ cell- specific gene 1 protein) (GSG1). | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.052585 (rank : 88) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ASB14_MOUSE
|
||||||
| NC score | 0.052486 (rank : 89) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.052386 (rank : 90) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ASB15_MOUSE
|
||||||
| NC score | 0.052096 (rank : 91) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.052041 (rank : 92) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
OSBL1_HUMAN
|
||||||
| NC score | 0.052000 (rank : 93) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
CDN2D_HUMAN
|
||||||
| NC score | 0.051765 (rank : 94) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
ASB13_HUMAN
|
||||||
| NC score | 0.051760 (rank : 95) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXK3, Q96EP7, Q9H8Z1 | Gene names | ASB13 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 13 (ASB-13). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.051681 (rank : 96) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
ANKR2_HUMAN
|
||||||
| NC score | 0.051599 (rank : 97) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9GZV1, Q5T456, Q8WUD7 | Gene names | ANKRD2, ARPP | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (hArpp). | |||||
|
ANR16_HUMAN
|
||||||
| NC score | 0.051498 (rank : 98) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P6B7, Q9NT01 | Gene names | ANKRD16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 16. | |||||
|
ASB9_MOUSE
|
||||||
| NC score | 0.051434 (rank : 99) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT8, Q9DAF7 | Gene names | Asb9 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
GABP2_HUMAN
|
||||||
| NC score | 0.051274 (rank : 100) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q06547, Q06545, Q12940, Q12941, Q12942, Q8IYD0 | Gene names | GABPB2, E4TF1B, GABPB, GABPB1 | |||
|
Domain Architecture |

|
|||||
| Description | GA-binding protein beta chain (GABP subunit beta-2) (GABP-2) (GABP subunit beta-1) (GABPB-1) (Transcription factor E4TF1-53) (Transcription factor E4TF1-47) (Nuclear respiratory factor 2). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.051162 (rank : 101) | θ value | 0.163984 (rank : 16) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.050883 (rank : 102) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
ZIM10_HUMAN
|
||||||
| NC score | 0.050780 (rank : 103) | θ value | 0.00390308 (rank : 12) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9ULJ6, Q7Z7E6 | Gene names | RAI17, KIAA1224, ZIMP10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
RFXK_HUMAN
|
||||||
| NC score | 0.050528 (rank : 104) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O14593, O95839 | Gene names | RFXANK, ANKRA1, RFXB | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein RFXANK (Regulatory factor X subunit B) (RFX-B) (Ankyrin repeat family A protein 1). | |||||
|
ASB13_MOUSE
|
||||||
| NC score | 0.050362 (rank : 105) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBX0 | Gene names | Asb13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 13 (ASB-13). | |||||
|
ANR29_HUMAN
|
||||||
| NC score | 0.050116 (rank : 106) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N6D5, Q6ZWE8, Q96LU9 | Gene names | ANKRD29 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 29. | |||||
|
ANKR2_MOUSE
|
||||||
| NC score | 0.050055 (rank : 107) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV06 | Gene names | Ankrd2, Arpp | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 2 (Skeletal muscle ankyrin repeat protein) (mArpp). | |||||
|
ANR25_HUMAN
|
||||||
| NC score | 0.050047 (rank : 108) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63ZY3, Q3KQZ3, Q6GUF5, Q9H8S4, Q9NUP0, Q9P210 | Gene names | ANKRD25, KIAA1518, SIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25 (SRC-1-interacting protein) (SRC1-interacting protein). | |||||
|
MANS1_HUMAN
|
||||||
| NC score | 0.045773 (rank : 109) | θ value | 0.163984 (rank : 18) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H8J5, Q8NEC1 | Gene names | MANSC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MANSC domain-containing protein 1 precursor. | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.044984 (rank : 110) | θ value | 0.21417 (rank : 19) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
ZIM10_MOUSE
|
||||||
| NC score | 0.043449 (rank : 111) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q6P1E1, Q6PDK9, Q6PF85, Q8BW47 | Gene names | Rai17, Zimp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 17 (PIAS-like protein Zimp10). | |||||
|
BAT2_MOUSE
|
||||||
| NC score | 0.035994 (rank : 112) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q7TSC1, Q923A9, Q9Z1R1 | Gene names | Bat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ZIMP7_MOUSE
|
||||||
| NC score | 0.035227 (rank : 113) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 566 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8CIE2, Q6P558, Q6P9Q5, Q8BJ00 | Gene names | Zimp7, D11Bwg0280e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PIAS-like protein Zimp7. | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.035114 (rank : 114) | θ value | 0.47712 (rank : 24) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.035081 (rank : 115) | θ value | 0.47712 (rank : 23) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
IRS2_MOUSE
|
||||||
| NC score | 0.031842 (rank : 116) | θ value | 0.163984 (rank : 17) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P81122 | Gene names | Irs2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Insulin receptor substrate 2 (IRS-2) (4PS). | |||||
|
SIN3A_HUMAN
|
||||||
| NC score | 0.030014 (rank : 117) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96ST3, Q8N8N4, Q8NC83, Q8WV18, Q96L98, Q9UFQ1 | Gene names | SIN3A | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SIN3A_MOUSE
|
||||||
| NC score | 0.029683 (rank : 118) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q60520, Q60820, Q62139, Q62140, Q7TPU8, Q7TSZ2 | Gene names | Sin3a | |||
|
Domain Architecture |

|
|||||
| Description | Paired amphipathic helix protein Sin3a (Transcriptional corepressor Sin3a) (Histone deacetylase complex subunit Sin3a). | |||||
|
SYNJ1_HUMAN
|
||||||
| NC score | 0.029627 (rank : 119) | θ value | 0.125558 (rank : 15) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O43426, O43425, O94984 | Gene names | SYNJ1, KIAA0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CA051_MOUSE
|
||||||
| NC score | 0.029456 (rank : 120) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3TQ03, Q3U1B8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf51 homolog. | |||||
|
NFIB_MOUSE
|
||||||
| NC score | 0.028119 (rank : 121) | θ value | 0.365318 (rank : 20) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97863, P70252, P70253, P70254, Q9R1G4 | Gene names | Nfib | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.027889 (rank : 122) | θ value | 1.81305 (rank : 37) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
NFIB_HUMAN
|
||||||
| NC score | 0.027381 (rank : 123) | θ value | 0.813845 (rank : 27) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00712, O00166, Q12858, Q96J45 | Gene names | NFIB | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor 1 B-type (Nuclear factor 1/B) (NF1-B) (NFI-B) (NF-I/B) (CCAAT-box-binding transcription factor) (CTF) (TGGCA-binding protein). | |||||
|
SYNJ1_MOUSE
|
||||||
| NC score | 0.023801 (rank : 124) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8CHC4 | Gene names | Synj1, Kiaa0910 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptojanin-1 (EC 3.1.3.36) (Synaptic inositol-1,4,5-trisphosphate 5- phosphatase 1). | |||||
|
CD45_MOUSE
|
||||||
| NC score | 0.023545 (rank : 125) | θ value | 0.00298849 (rank : 11) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06800, Q61812, Q61813, Q61814, Q61815, Q78EF1 | Gene names | Ptprc, Ly-5 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte common antigen precursor (EC 3.1.3.48) (L-CA) (Lymphocyte common antigen Ly-5) (CD45 antigen) (T200). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.023492 (rank : 126) | θ value | 0.62314 (rank : 25) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PCGF2_MOUSE
|
||||||
| NC score | 0.023291 (rank : 127) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P23798 | Gene names | Pcgf2, Mel-18, Mel18, Rnf110, Zfp144, Znf144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144) (Zfp-144). | |||||
|
SOX7_MOUSE
|
||||||
| NC score | 0.022422 (rank : 128) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P40646, Q9R1T6 | Gene names | Sox7, Sox-7 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor SOX-7 (mSOX7). | |||||
|
MAVS_HUMAN
|
||||||
| NC score | 0.021926 (rank : 129) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z434, Q3I0Y2, Q5T7I6, Q86VY7, Q9H1H3, Q9H4Y1, Q9H8D3, Q9ULE9 | Gene names | MAVS, IPS1, KIAA1271, VISA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial antiviral signaling protein (Interferon-beta promoter stimulator protein 1) (IPS-1) (Virus-induced signaling adapter) (CARD adapter inducing interferon-beta) (Cardif) (Putative NF-kappa-B- activating protein 031N). | |||||
|
PCGF2_HUMAN
|
||||||
| NC score | 0.021610 (rank : 130) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35227 | Gene names | PCGF2, MEL18, RNF110, ZNF144 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb group RING finger protein 2 (DNA-binding protein Mel-18) (RING finger protein 110) (Zinc finger protein 144). | |||||
|
CLOCK_MOUSE
|
||||||
| NC score | 0.021055 (rank : 131) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08785 | Gene names | Clock | |||
|
Domain Architecture |

|
|||||
| Description | Circadian locomoter output cycles protein kaput (mCLOCK). | |||||
|
TM108_HUMAN
|
||||||
| NC score | 0.020861 (rank : 132) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UXF1, Q9BQH1, Q9BW81, Q9C0H3 | Gene names | TMEM108, KIAA1690 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.020325 (rank : 133) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.019961 (rank : 134) | θ value | 0.47712 (rank : 22) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PER3_HUMAN
|
||||||
| NC score | 0.019409 (rank : 135) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 479 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P56645, Q5H8X4, Q969K6, Q96S77, Q96S78, Q9C0J3, Q9NSP9, Q9UGU8 | Gene names | PER3 | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 3 (hPER3). | |||||
|
PRR8_HUMAN
|
||||||
| NC score | 0.018894 (rank : 136) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NSV0 | Gene names | PRR8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 8. | |||||
|
TEX2_HUMAN
|
||||||
| NC score | 0.018775 (rank : 137) | θ value | 2.36792 (rank : 43) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IWB9, Q6AHZ5, Q8N3L0, Q9C0C5 | Gene names | TEX2, KIAA1738 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 2 protein. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.018719 (rank : 138) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.018323 (rank : 139) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.017788 (rank : 140) | θ value | 1.06291 (rank : 30) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
GR65_MOUSE
|
||||||
| NC score | 0.017026 (rank : 141) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91X51, Q9D3L9 | Gene names | Gorasp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.016990 (rank : 142) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
PGFRL_MOUSE
|
||||||
| NC score | 0.016552 (rank : 143) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PE55, Q3V182, Q9D0Z1 | Gene names | Pdgfrl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Platelet-derived growth factor receptor-like protein precursor. | |||||
|
TTLL5_MOUSE
|
||||||
| NC score | 0.016439 (rank : 144) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8CHB8 | Gene names | Ttll5, Kiaa0998 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tubulin--tyrosine ligase-like protein 5. | |||||
|
MAML2_HUMAN
|
||||||
| NC score | 0.015465 (rank : 145) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8IZL2, Q6AI23, Q6Y3A3, Q8IUL3, Q96JK6 | Gene names | MAML2, KIAA1819 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 2 (Mam-2). | |||||
|
AKAP2_HUMAN
|
||||||
| NC score | 0.015333 (rank : 146) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Y2D5, Q9UG26 | Gene names | AKAP2, KIAA0920 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 2 (Protein kinase A-anchoring protein 2) (PRKA2) (AKAP-2). | |||||
|
CNOT4_MOUSE
|
||||||
| NC score | 0.013756 (rank : 147) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BT14, Q8CCR4, Q9CV74, Q9Z1D0 | Gene names | Cnot4, Not4 | |||
|
Domain Architecture |

|
|||||
| Description | CCR4-NOT transcription complex subunit 4 (EC 6.3.2.-) (E3 ubiquitin protein ligase CNOT4) (CCR4-associated factor 4) (Potential transcriptional repressor NOT4Hp). | |||||
|
ATX7_HUMAN
|
||||||
| NC score | 0.013683 (rank : 148) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15265, O75328, O75329, Q9Y6P8 | Gene names | ATXN7, SCA7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-7 (Spinocerebellar ataxia type 7 protein). | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.013218 (rank : 149) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
VP13A_HUMAN
|
||||||
| NC score | 0.013124 (rank : 150) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96RL7, Q5JSX9, Q5JSY0, Q5VYR5, Q702P4, Q709D0, Q86YF8, Q96S61, Q9H995, Q9Y2J1 | Gene names | VPS13A, CHAC, KIAA0986 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 13A (Chorein) (Chorea- acanthocytosis protein). | |||||
|
OLIG2_HUMAN
|
||||||
| NC score | 0.012511 (rank : 151) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q13516, Q86X04, Q9NZ14 | Gene names | OLIG2, BHLHB1, PRKCBP2, RACK17 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2) (Basic helix-loop- helix protein class B 1) (Protein kinase C-binding protein RACK17) (Protein kinase C-binding protein 2). | |||||
|
MINT_HUMAN
|
||||||
| NC score | 0.011616 (rank : 152) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
ODO2_HUMAN
|
||||||
| NC score | 0.011423 (rank : 153) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36957, Q9BQ32 | Gene names | DLST, DLTS | |||
|
Domain Architecture |

|
|||||
| Description | Dihydrolipoyllysine-residue succinyltransferase component of 2- oxoglutarate dehydrogenase complex, mitochondrial precursor (EC 2.3.1.61) (Dihydrolipoamide succinyltransferase component of 2- oxoglutarate dehydrogenase complex) (E2) (E2K). | |||||
|
CAC1C_HUMAN
|
||||||
| NC score | 0.010631 (rank : 154) | θ value | 0.47712 (rank : 21) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13936, Q13917, Q13918, Q13919, Q13920, Q13921, Q13922, Q13923, Q13924, Q13925, Q13926, Q13927, Q13928, Q13929, Q13930, Q13932, Q13933, Q14743, Q14744, Q15877, Q99025, Q99241, Q99875 | Gene names | CACNA1C, CACH2, CACN2, CACNL1A1, CCHL1A1 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent L-type calcium channel subunit alpha-1C (Voltage- gated calcium channel subunit alpha Cav1.2) (Calcium channel, L type, alpha-1 polypeptide, isoform 1, cardiac muscle). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.010534 (rank : 155) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
DPPA2_MOUSE
|
||||||
| NC score | 0.010238 (rank : 156) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CWH0 | Gene names | Dppa2, Phsecrg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Developmental pluripotency-associated protein 2 (Pluripotent embryonic stem cell-related protein 1). | |||||
|
OLIG2_MOUSE
|
||||||
| NC score | 0.009962 (rank : 157) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 74 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9EQW6, Q9JKN4 | Gene names | Olig2 | |||
|
Domain Architecture |

|
|||||
| Description | Oligodendrocyte transcription factor 2 (Oligo2). | |||||
|
WBP1_MOUSE
|
||||||
| NC score | 0.009772 (rank : 158) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97764, Q8WUP5, Q99J20 | Gene names | Wbp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 1 (WBP-1). | |||||
|
GGA3_HUMAN
|
||||||
| NC score | 0.009625 (rank : 159) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZ52, Q15017, Q6IS16, Q9UJY3 | Gene names | GGA3, KIAA0154 | |||
|
Domain Architecture |

|
|||||
| Description | ADP-ribosylation factor-binding protein GGA3 (Golgi-localized, gamma ear-containing, ARF-binding protein 3). | |||||
|
IF4G3_HUMAN
|
||||||
| NC score | 0.009615 (rank : 160) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43432, Q15597, Q8NEN1 | Gene names | EIF4G3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 3 (eIF-4-gamma 3) (eIF-4G 3) (eIF4G 3) (eIF-4-gamma II) (eIF4GII). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.009228 (rank : 161) | θ value | 2.36792 (rank : 44) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
PTPRN_HUMAN
|
||||||
| NC score | 0.009194 (rank : 162) | θ value | 5.27518 (rank : 63) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16849, Q08319 | Gene names | PTPRN, ICA3, ICA512 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase-like N precursor (R-PTP-N) (PTP IA-2) (Islet cell antigen 512) (ICA 512) (Islet cell autoantigen 3). | |||||
|
AOC2_HUMAN
|
||||||
| NC score | 0.008449 (rank : 163) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75106, O00120, O75105, Q4TTW5, Q9UNY0 | Gene names | AOC2 | |||
|
Domain Architecture |

|
|||||
| Description | Retina-specific copper amine oxidase precursor (EC 1.4.3.6) (RAO) (Amine oxidase [copper-containing]). | |||||
|
UBQL2_HUMAN
|
||||||
| NC score | 0.008114 (rank : 164) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UHD9, O94798, Q9H3W6, Q9HAZ4 | Gene names | UBQLN2, PLIC2 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquilin-2 (Protein linking IAP with cytoskeleton 2) (PLIC-2) (hPLIC- 2) (Ubiquitin-like product Chap1/Dsk2) (DSK2 homolog) (Chap1). | |||||
|
FOXI1_HUMAN
|
||||||
| NC score | 0.008030 (rank : 165) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12951, Q14518, Q66SR7, Q8N6L8 | Gene names | FOXI1, FKHL10, FREAC6 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein I1 (Forkhead-related protein FKHL10) (Forkhead- related transcription factor 6) (FREAC-6) (Hepatocyte nuclear factor 3 forkhead homolog 3) (HNF-3/fork-head homolog 3) (HFH-3). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.007836 (rank : 166) | θ value | 0.0961366 (rank : 14) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
FOXI1_MOUSE
|
||||||
| NC score | 0.007821 (rank : 167) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q922I5, Q5SRI5, Q9D299 | Gene names | Foxi1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein I1. | |||||
|
AT10B_HUMAN
|
||||||
| NC score | 0.005797 (rank : 168) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94823, Q9H725 | Gene names | ATP10B, ATPVB, KIAA0715 | |||
|
Domain Architecture |

|
|||||
| Description | Probable phospholipid-transporting ATPase VB (EC 3.6.3.1). | |||||
|
NID1_MOUSE
|
||||||
| NC score | 0.003678 (rank : 169) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P10493, Q8BQI3, Q8C3U8, Q8C9P6 | Gene names | Nid1, Ent | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-1 precursor (Entactin). | |||||
|
LAP2_MOUSE
|
||||||
| NC score | 0.003289 (rank : 170) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 482 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80TH2, Q8BQ14, Q8CE41, Q8K171, Q99JU3, Q9JI47 | Gene names | Erbb2ip, Erbin, Kiaa1225, Lap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein LAP2 (Erbb2-interacting protein) (Erbin) (Densin-180-like protein). | |||||
|
PRDM2_HUMAN
|
||||||
| NC score | 0.000668 (rank : 171) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13029, Q13149, Q14550, Q5VUL9 | Gene names | PRDM2, RIZ | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 2 (PR domain-containing protein 2) (Retinoblastoma protein-interacting zinc-finger protein) (Zinc finger protein RIZ) (MTE-binding protein) (MTB-ZF) (GATA-3-binding protein G3B). | |||||
|
ULK2_HUMAN
|
||||||
| NC score | -0.000433 (rank : 172) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 77 | Target Neighborhood Hits | 979 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IYT8, O75119 | Gene names | ULK2, KIAA0623 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK2 (EC 2.7.11.1) (Unc-51-like kinase 2). | |||||