Please be patient as the page loads
|
TF65_MOUSE
|
||||||
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TF65_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985470 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
REL_HUMAN
|
||||||
| θ value | 8.73085e-104 (rank : 3) | NC score | 0.944896 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
REL_MOUSE
|
||||||
| θ value | 4.05562e-101 (rank : 4) | NC score | 0.939917 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
RELB_MOUSE
|
||||||
| θ value | 4.81303e-86 (rank : 5) | NC score | 0.946059 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |
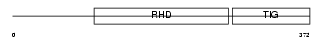
|
|||||
| Description | Transcription factor RelB. | |||||
|
RELB_HUMAN
|
||||||
| θ value | 5.32145e-85 (rank : 6) | NC score | 0.944721 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |
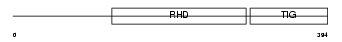
|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
NFKB2_HUMAN
|
||||||
| θ value | 4.68348e-65 (rank : 7) | NC score | 0.555748 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_MOUSE
|
||||||
| θ value | 6.11685e-65 (rank : 8) | NC score | 0.573394 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_MOUSE
|
||||||
| θ value | 5.35859e-61 (rank : 9) | NC score | 0.543430 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25799, Q3TZE8, Q3V2V6, Q6TDG8, Q75ZL1, Q80Y21, Q8C712 | Gene names | Nfkb1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) (NF-kappa-B1 p84/NF-kappa-B1 p98) [Contains: Nuclear factor NF- kappa-B p50 subunit]. | |||||
|
NFKB1_HUMAN
|
||||||
| θ value | 4.53632e-60 (rank : 10) | NC score | 0.542698 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
NFAC3_MOUSE
|
||||||
| θ value | 1.17247e-07 (rank : 11) | NC score | 0.253484 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_HUMAN
|
||||||
| θ value | 2.61198e-07 (rank : 12) | NC score | 0.250008 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_HUMAN
|
||||||
| θ value | 1.69304e-06 (rank : 13) | NC score | 0.221579 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAT5_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 14) | NC score | 0.298911 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFAT5_MOUSE
|
||||||
| θ value | 1.09739e-05 (rank : 15) | NC score | 0.306021 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAC1_MOUSE
|
||||||
| θ value | 1.43324e-05 (rank : 16) | NC score | 0.215696 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC2_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 17) | NC score | 0.200722 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.010287 (rank : 129) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 0.47712 (rank : 19) | NC score | 0.028135 (rank : 108) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.037626 (rank : 99) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.023196 (rank : 115) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.026445 (rank : 113) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
KLF17_HUMAN
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.001994 (rank : 142) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
MLL2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.020680 (rank : 117) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TAB3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.020308 (rank : 118) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
IL7RA_MOUSE
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.036780 (rank : 100) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16872 | Gene names | Il7r | |||
|
Domain Architecture |
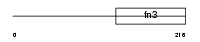
|
|||||
| Description | Interleukin-7 receptor alpha chain precursor (IL-7R-alpha) (CD127 antigen). | |||||
|
RAI2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.032379 (rank : 102) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
GAK11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.028549 (rank : 107) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.028836 (rank : 104) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.028659 (rank : 105) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.028555 (rank : 106) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK6_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.029117 (rank : 103) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GNAS3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.040487 (rank : 98) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
K1958_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.018496 (rank : 119) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
PARC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.013195 (rank : 127) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
WEE1B_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.003015 (rank : 141) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
BL1S3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.025110 (rank : 114) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6QNY0 | Gene names | BLOC1S3, BLOS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Biogenesis of lysosome-related organelles complex-1 subunit 3 (BLOC subunit 3). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.022213 (rank : 116) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
IPMK_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.015462 (rank : 124) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFU5 | Gene names | IPMK, IMPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol polyphosphate multikinase (EC 2.7.1.151) (Inositol 1,3,4,6- tetrakisphosphate 5-kinase). | |||||
|
TNKS2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.060971 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
LPP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.016085 (rank : 122) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
C8AP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.014095 (rank : 126) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
GAK1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.027786 (rank : 109) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
KTNB1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.001850 (rank : 143) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG40, Q8CD18, Q8R1J0, Q9CWV2 | Gene names | Katnb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p80 WD40-containing subunit B1 (Katanin p80 subunit B1) (p80 katanin). | |||||
|
NKX23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.012212 (rank : 128) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TAU0, Q9NYS6 | Gene names | NKX2-3, NKX23, NKX2C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.3 (Homeobox protein NKX-2.3) (Homeobox protein NK-2 homolog C). | |||||
|
NKX24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.008120 (rank : 135) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2Z4 | Gene names | NKX2-4, NKX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.4 (Homeobox protein NKX2.4) (Homeobox protein NK-2 homolog D). | |||||
|
PTN23_HUMAN
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.007840 (rank : 136) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
TARA_MOUSE
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.006365 (rank : 138) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
YLPM1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.026641 (rank : 112) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.033422 (rank : 101) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.018208 (rank : 120) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.009984 (rank : 131) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.016876 (rank : 121) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
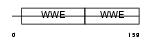
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
GAK5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.027485 (rank : 110) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
SELPL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.014343 (rank : 125) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.005673 (rank : 139) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
AAKG2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.009207 (rank : 133) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
ACRO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.001389 (rank : 144) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |
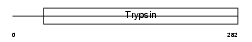
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
CASK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.016064 (rank : 123) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07498, Q13575 | Gene names | CSN3, CASK, CSN10, CSNK | |||
|
Domain Architecture |

|
|||||
| Description | Kappa-casein precursor. | |||||
|
GAK3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.027087 (rank : 111) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
LTBP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.001267 (rank : 145) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||
|
MNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.010069 (rank : 130) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
PAR3L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.004571 (rank : 140) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
PPCKC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.006463 (rank : 137) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2V4, Q3UEH3 | Gene names | Pck1, Pepck | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoenolpyruvate carboxykinase, cytosolic [GTP] (EC 4.1.1.32) (Phosphoenolpyruvate carboxylase) (PEPCK-C). | |||||
|
RNF37_MOUSE
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.008516 (rank : 134) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925F4 | Gene names | Ubox5, Rnf37, Ubce7ip5, Uip5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (UbcM4-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
TDG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.009930 (rank : 132) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56581 | Gene names | Tdg | |||
|
Domain Architecture |

|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-) (C-JUN leucine zipper interactive protein JZA-3). | |||||
|
ABTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.051091 (rank : 90) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
ANFY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.055450 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANFY1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.053838 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.054350 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.053663 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.051927 (rank : 87) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ANK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.054340 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
ANKR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.053302 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANKR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.053956 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANKR6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.063894 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
ANKR7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.057682 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |
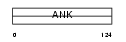
|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ANR10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.058673 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |
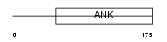
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.056684 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |
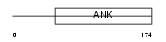
|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR23_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.057821 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANR23_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.054377 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANR25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.051421 (rank : 89) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ANR28_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.053022 (rank : 82) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.053260 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR33_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056072 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ANR41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.053580 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
ANR42_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.067216 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR42_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053995 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANR44_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.052894 (rank : 83) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
ANR50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.050464 (rank : 94) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
ANR52_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.056628 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANR52_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.057807 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ANRA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.055806 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ANRA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.054963 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
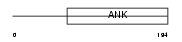
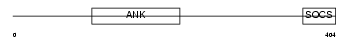
ASB10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.051466 (rank : 88) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.056920 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ASB14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.050682 (rank : 92) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
ASB15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.050025 (rank : 97) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
ASB16_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.055786 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |
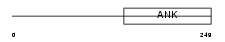
|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.066098 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ASB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.058815 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.059697 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.052273 (rank : 85) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWX0 | Gene names | ASB5 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 5 (ASB-5). | |||||
|
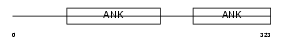
ASB6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.061725 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.069173 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |
|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
ASB7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.056108 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |
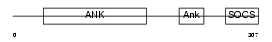
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.056004 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |
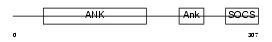
|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ASB8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.054552 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |
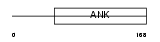
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.054000 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |
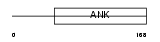
|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ASB9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.054845 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DX5, Q9NWS5, Q9Y4T3 | Gene names | ASB9 | |||
|
Domain Architecture |
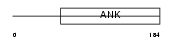
|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
BCL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.112128 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |
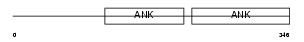
|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
BCL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.112817 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
CDN2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.050193 (rank : 96) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |
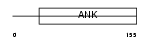
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CDN2D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.052093 (rank : 86) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |
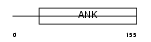
|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
CSKI1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.054091 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.054232 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
FANK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.050481 (rank : 93) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAM9, Q9CUA7 | Gene names | Fank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains 1 protein (Germ cell- specific gene 1 protein) (GSG1). | |||||
|
IKBA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.125855 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |
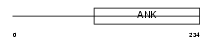
|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.126598 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.127563 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
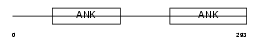
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.125256 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |
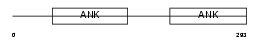
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
IKBE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.135728 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
IKBE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.129564 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
IKBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.072852 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |
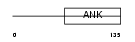
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
IKBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.071009 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |
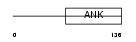
|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
INVS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.051081 (rank : 91) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
MIB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.066198 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
MIB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.066268 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.060390 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
MIB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.056855 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
NFAC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.149798 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.176781 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
OSBL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.050217 (rank : 95) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
PA2G6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.067537 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PA2G6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.063248 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
PSD10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.065761 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |
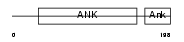
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.065586 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |
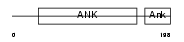
|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
SNCAP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.065069 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
TNKS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.058786 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
TRPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.052362 (rank : 84) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
TRPA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.053473 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ZDH13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.055001 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |
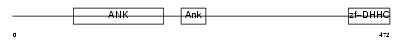
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ZDH13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.054582 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |
|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ZDH17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.057695 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ZDH17_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.056769 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |
|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.985470 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
RELB_MOUSE
|
||||||
| NC score | 0.946059 (rank : 3) | θ value | 4.81303e-86 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q04863 | Gene names | Relb | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB. | |||||
|
REL_HUMAN
|
||||||
| NC score | 0.944896 (rank : 4) | θ value | 8.73085e-104 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q04864, Q2PNZ7, Q6LDY0 | Gene names | REL | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
RELB_HUMAN
|
||||||
| NC score | 0.944721 (rank : 5) | θ value | 5.32145e-85 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q01201, Q6GTX7, Q9UEI7 | Gene names | RELB | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor RelB (I-Rel). | |||||
|
REL_MOUSE
|
||||||
| NC score | 0.939917 (rank : 6) | θ value | 4.05562e-101 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15307 | Gene names | Rel | |||
|
Domain Architecture |

|
|||||
| Description | C-Rel proto-oncogene protein (C-Rel protein). | |||||
|
NFKB2_MOUSE
|
||||||
| NC score | 0.573394 (rank : 7) | θ value | 6.11685e-65 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9WTK5 | Gene names | Nfkb2 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB2_HUMAN
|
||||||
| NC score | 0.555748 (rank : 8) | θ value | 4.68348e-65 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q00653, Q04860, Q9BU75, Q9H471, Q9H472 | Gene names | NFKB2, LYT10 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p100 subunit (DNA-binding factor KBF2) (H2TF1) (Lymphocyte translocation chromosome 10) (Oncogene Lyt-10) (Lyt10) [Contains: Nuclear factor NF-kappa-B p52 subunit]. | |||||
|
NFKB1_MOUSE
|
||||||
| NC score | 0.543430 (rank : 9) | θ value | 5.35859e-61 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P25799, Q3TZE8, Q3V2V6, Q6TDG8, Q75ZL1, Q80Y21, Q8C712 | Gene names | Nfkb1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) (NF-kappa-B1 p84/NF-kappa-B1 p98) [Contains: Nuclear factor NF- kappa-B p50 subunit]. | |||||
|
NFKB1_HUMAN
|
||||||
| NC score | 0.542698 (rank : 10) | θ value | 4.53632e-60 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P19838, Q68D84, Q86V43, Q8N4X7, Q9NZC0 | Gene names | NFKB1 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor NF-kappa-B p105 subunit (DNA-binding factor KBF1) (EBP- 1) [Contains: Nuclear factor NF-kappa-B p50 subunit]. | |||||
|
NFAT5_MOUSE
|
||||||
| NC score | 0.306021 (rank : 11) | θ value | 1.09739e-05 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9WV30 | Gene names | Nfat5 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T cell transcription factor NFAT5) (NF-AT5) (Rel domain-containing transcription factor NFAT5). | |||||
|
NFAT5_HUMAN
|
||||||
| NC score | 0.298911 (rank : 12) | θ value | 1.09739e-05 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O94916, O95693, Q9UN18 | Gene names | NFAT5, KIAA0827, TONEBP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells 5 (T-cell transcription factor NFAT5) (NF-AT5) (Tonicity-responsive enhancer-binding protein) (TonE- binding protein) (TonEBP). | |||||
|
NFAC3_MOUSE
|
||||||
| NC score | 0.253484 (rank : 13) | θ value | 1.17247e-07 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P97305, Q60896 | Gene names | Nfatc3, Nfat4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC3_HUMAN
|
||||||
| NC score | 0.250008 (rank : 14) | θ value | 2.61198e-07 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q12968, O75211, Q14516, Q99840, Q99841, Q99842 | Gene names | NFATC3, NFAT4 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 3 (NF-ATc3) (NFATc3) (T cell transcription factor NFAT4) (NF-AT4) (NFATx). | |||||
|
NFAC1_HUMAN
|
||||||
| NC score | 0.221579 (rank : 15) | θ value | 1.69304e-06 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | O95644, Q12865, Q15793 | Gene names | NFATC1, NFAT2, NFATC | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC1_MOUSE
|
||||||
| NC score | 0.215696 (rank : 16) | θ value | 1.43324e-05 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O88942, O70345 | Gene names | Nfatc1, Nfat2, Nfatc | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 1 (NFAT transcription complex cytosolic component) (NF-ATc1) (NF-ATc). | |||||
|
NFAC2_MOUSE
|
||||||
| NC score | 0.200722 (rank : 17) | θ value | 5.44631e-05 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 98 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q60591, Q60984, Q60985 | Gene names | Nfatc2, Nfat1, Nfatp | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
NFAC4_HUMAN
|
||||||
| NC score | 0.176781 (rank : 18) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14934 | Gene names | NFATC4, NFAT3 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 4 (NF-ATc4) (NFATc4) (T cell transcription factor NFAT3) (NF-AT3). | |||||
|
NFAC2_HUMAN
|
||||||
| NC score | 0.149798 (rank : 19) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q13469, Q13468, Q5TFW7, Q5TFW8, Q9NPX6, Q9NQH3, Q9UJR2 | Gene names | NFATC2, NFAT1, NFATP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear factor of activated T-cells, cytoplasmic 2 (T cell transcription factor NFAT1) (NFAT pre-existing subunit) (NF-ATp). | |||||
|
IKBE_HUMAN
|
||||||
| NC score | 0.135728 (rank : 20) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00221 | Gene names | NFKBIE, IKBE | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKBE). | |||||
|
IKBE_MOUSE
|
||||||
| NC score | 0.129564 (rank : 21) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O54910, Q3U686, Q9CZZ9, Q9D7U3 | Gene names | Nfkbie, Ikbe | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor epsilon (NF-kappa-BIE) (I-kappa-B-epsilon) (IkappaBepsilon) (IKB-epsilon) (IKB-E). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.127563 (rank : 22) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
IKBA_MOUSE
|
||||||
| NC score | 0.126598 (rank : 23) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z1E3, Q80ZX5 | Gene names | Nfkbia, Ikba | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B inhibitor alpha (I-kappa-B-alpha) (IkappaBalpha) (IKB- alpha). | |||||
|
IKBA_HUMAN
|
||||||
| NC score | 0.125855 (rank : 24) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25963 | Gene names | NFKBIA, IKBA, MAD3, NFKBI | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor alpha (Major histocompatibility complex enhancer- binding protein MAD3) (I-kappa-B-alpha) (IkappaBalpha) (IKB-alpha). | |||||
|
IKBB_MOUSE
|
||||||
| NC score | 0.125256 (rank : 25) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60778, Q9D6L5 | Gene names | Nfkbib, Ikbb | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B). | |||||
|
BCL3_MOUSE
|
||||||
| NC score | 0.112817 (rank : 26) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z2F6 | Gene names | Bcl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 3-encoded protein homolog (Protein Bcl-3). | |||||
|
BCL3_HUMAN
|
||||||
| NC score | 0.112128 (rank : 27) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20749 | Gene names | BCL3 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell lymphoma 3-encoded protein (Protein Bcl-3). | |||||
|
IKBL_HUMAN
|
||||||
| NC score | 0.072852 (rank : 28) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UBC1, Q14625, Q9UBX4 | Gene names | NFKBIL1, IKBL | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
IKBL_MOUSE
|
||||||
| NC score | 0.071009 (rank : 29) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88995, Q91X16, Q9R1Y3 | Gene names | Nfkbil1, Ikbl | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor-like protein 1 (Inhibitor of kappa B-like) (I- kappa-B-like) (IkappaBL) (Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1). | |||||
|
ASB6_MOUSE
|
||||||
| NC score | 0.069173 (rank : 30) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU1, Q8VEC6 | Gene names | Asb6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
PA2G6_HUMAN
|
||||||
| NC score | 0.067537 (rank : 31) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60733, O75645, Q8N452, Q9UG29, Q9UIT0, Q9Y671 | Gene names | PLA2G6, IPLA2 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ANR42_HUMAN
|
||||||
| NC score | 0.067216 (rank : 32) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N9B4, Q49A49 | Gene names | ANKRD42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
MIB1_MOUSE
|
||||||
| NC score | 0.066268 (rank : 33) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q80SY4, Q5XK51, Q6IS57, Q6YI52, Q6ZPT8, Q8BNR1, Q8C6W2, Q921Q1 | Gene names | Mib1, Dip1, Kiaa1323, Mib | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1). | |||||
|
MIB1_HUMAN
|
||||||
| NC score | 0.066198 (rank : 34) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 470 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86YT6, Q68D01, Q6YI51, Q8NBY0, Q8TCB5, Q8TCL7, Q9P2M3 | Gene names | MIB1, DIP1, KIAA1323, ZZANK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB1 (EC 6.3.2.-) (Mind bomb homolog 1) (DAPK-interacting protein 1) (DIP-1) (Zinc finger ZZ type with ankyrin repeat domain protein 2). | |||||
|
ASB16_MOUSE
|
||||||
| NC score | 0.066098 (rank : 35) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 306 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS5, Q8BYT0 | Gene names | Asb16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
PSD10_HUMAN
|
||||||
| NC score | 0.065761 (rank : 36) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75832 | Gene names | PSMD10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
PSD10_MOUSE
|
||||||
| NC score | 0.065586 (rank : 37) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z2X2, Q9D7N8 | Gene names | Psmd10 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 10 (26S proteasome regulatory subunit p28) (Gankyrin). | |||||
|
SNCAP_HUMAN
|
||||||
| NC score | 0.065069 (rank : 38) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y6H5 | Gene names | SNCAIP | |||
|
Domain Architecture |

|
|||||
| Description | Synphilin-1 (Alpha-synuclein-interacting protein). | |||||
|
ANKR6_HUMAN
|
||||||
| NC score | 0.063894 (rank : 39) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2G4, Q9NU24, Q9UFQ9 | Gene names | ANKRD6, KIAA0957 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 6. | |||||
|
PA2G6_MOUSE
|
||||||
| NC score | 0.063248 (rank : 40) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97819, Q99LA9, Q9JK61 | Gene names | Pla2g6 | |||
|
Domain Architecture |

|
|||||
| Description | 85 kDa calcium-independent phospholipase A2 (EC 3.1.1.4) (iPLA2) (CaI- PLA2) (Group VI phospholipase A2) (GVI PLA2). | |||||
|
ASB6_HUMAN
|
||||||
| NC score | 0.061725 (rank : 41) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWX5 | Gene names | ASB6 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 6 (ASB-6). | |||||
|
TNKS2_HUMAN
|
||||||
| NC score | 0.060971 (rank : 42) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2K2, Q9H8F2, Q9HAS4 | Gene names | TNKS2, PARP5B, TANK2, TNKL | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-2 (EC 2.4.2.30) (TANK2) (Tankyrase II) (TNKS-2) (TRF1- interacting ankyrin-related ADP-ribose polymerase 2) (Tankyrase-like protein) (Tankyrase-related protein). | |||||
|
MIB2_HUMAN
|
||||||
| NC score | 0.060390 (rank : 43) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AX9, Q7Z437, Q8IY62, Q8N897, Q8N8R2, Q8N911, Q8NB36, Q8NCY1, Q8NG59, Q8NG60, Q8NG61, Q8NI59, Q8WYN1 | Gene names | MIB2, SKD, ZZANK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Zinc finger ZZ type with ankyrin repeat domain protein 1) (Skeletrophin) (Novelzin) (Novel zinc finger protein) (Putative NF-kappa-B-activating protein 002N). | |||||
|
ASB3_MOUSE
|
||||||
| NC score | 0.059697 (rank : 44) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9WV72 | Gene names | Asb3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
ASB3_HUMAN
|
||||||
| NC score | 0.058815 (rank : 45) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y575, Q9NVZ2 | Gene names | ASB3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 3 (ASB-3). | |||||
|
TNKS1_HUMAN
|
||||||
| NC score | 0.058786 (rank : 46) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 477 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95271, O95272 | Gene names | TNKS, PARP5A, PARPL, TIN1, TINF1, TNKS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tankyrase-1 (EC 2.4.2.30) (TANK1) (Tankyrase I) (TNKS-1) (TRF1- interacting ankyrin-related ADP-ribose polymerase). | |||||
|
ANR10_HUMAN
|
||||||
| NC score | 0.058673 (rank : 47) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NXR5 | Gene names | ANKRD10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR23_HUMAN
|
||||||
| NC score | 0.057821 (rank : 48) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86SG2, Q711K7, Q8NAJ7 | Gene names | ANKRD23, DARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein) (Muscle ankyrin repeat protein 3). | |||||
|
ANR52_MOUSE
|
||||||
| NC score | 0.057807 (rank : 49) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BTI7 | Gene names | Ankrd52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ZDH17_HUMAN
|
||||||
| NC score | 0.057695 (rank : 50) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH5, O75407, Q7Z2I0, Q86W89, Q86YK0, Q9P088, Q9UPZ8 | Gene names | ZDHHC17, HIP14, HIP3, KIAA0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14) (Huntingtin-interacting protein 3) (Huntingtin-interacting protein HYPH) (Putative NF-kappa-B-activating protein 205) (Putative MAPK- activating protein PM11). | |||||
|
ANKR7_HUMAN
|
||||||
| NC score | 0.057682 (rank : 51) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92527, Q96QN1, Q9UDM3 | Gene names | ANKRD7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 7 (Testis-specific protein TSA806). | |||||
|
ASB10_MOUSE
|
||||||
| NC score | 0.056920 (rank : 52) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 301 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT7 | Gene names | Asb10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
MIB2_MOUSE
|
||||||
| NC score | 0.056855 (rank : 53) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8R516, Q52QU8, Q6PEF6, Q8C1N7, Q8VIB4 | Gene names | Mib2, Skd | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein MIB2 (EC 6.3.2.-) (Mind bomb homolog 2) (Mind bomb-2) (Skeletrophin) (Dystrophin-like protein) (Dyslike). | |||||
|
ZDH17_MOUSE
|
||||||
| NC score | 0.056769 (rank : 54) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 328 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TN5, Q8BJ08, Q8BYQ2, Q8BYQ6 | Gene names | Zdhhc17, Hip14, Kiaa0946 | |||
|
Domain Architecture |

|
|||||
| Description | Palmitoyltransferase ZDHHC17 (EC 2.3.1.-) (Zinc finger DHHC domain- containing protein 17) (DHHC-17) (Huntingtin-interacting protein 14). | |||||
|
ANR10_MOUSE
|
||||||
| NC score | 0.056684 (rank : 55) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99LW0 | Gene names | Ankrd10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 10. | |||||
|
ANR52_HUMAN
|
||||||
| NC score | 0.056628 (rank : 56) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NB46 | Gene names | ANKRD52 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 52. | |||||
|
ASB7_HUMAN
|
||||||
| NC score | 0.056108 (rank : 57) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H672, Q7Z4S3 | Gene names | ASB7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ANR33_HUMAN
|
||||||
| NC score | 0.056072 (rank : 58) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q7Z3H0, Q5K619, Q5K621, Q5K622, Q5K623, Q5K624, Q6ZUN0 | Gene names | ANKRD33, C12orf7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 33. | |||||
|
ASB7_MOUSE
|
||||||
| NC score | 0.056004 (rank : 59) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU0 | Gene names | Asb7 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 7 (ASB-7). | |||||
|
ANRA2_HUMAN
|
||||||
| NC score | 0.055806 (rank : 60) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H9E1 | Gene names | ANKRA2, ANKRA | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB16_HUMAN
|
||||||
| NC score | 0.055786 (rank : 61) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96NS5, Q8WXK0 | Gene names | ASB16 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 16 (ASB-16). | |||||
|
ANFY1_HUMAN
|
||||||
| NC score | 0.055450 (rank : 62) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9P2R3, Q9ULG5 | Gene names | ANKFY1, ANKHZN, KIAA1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ZDH13_HUMAN
|
||||||
| NC score | 0.055001 (rank : 63) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IUH4, Q7Z2D3, Q86VK2, Q9NV30, Q9NV99 | Gene names | ZDHHC13, HIP14L, HIP3RP | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein) (Huntingtin- interacting protein HIP3RP) (Putative NF-kappa-B-activating protein 209) (Putative MAPK-activating protein PM03). | |||||
|
ANRA2_MOUSE
|
||||||
| NC score | 0.054963 (rank : 64) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99PE2 | Gene names | Ankra2, Ankra | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat family A protein 2 (RFXANK-like 2). | |||||
|
ASB9_HUMAN
|
||||||
| NC score | 0.054845 (rank : 65) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96DX5, Q9NWS5, Q9Y4T3 | Gene names | ASB9 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 9 (ASB-9). | |||||
|
ZDH13_MOUSE
|
||||||
| NC score | 0.054582 (rank : 66) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CWU2, Q3UK32, Q3UKV1 | Gene names | Zdhhc13, Hip14l | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC13 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 13) (DHHC-13) (Huntingtin-interacting protein 14-related protein) (HIP14-related protein). | |||||
|
ASB8_HUMAN
|
||||||
| NC score | 0.054552 (rank : 67) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9H765, Q547Q2 | Gene names | ASB8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ANR23_MOUSE
|
||||||
| NC score | 0.054377 (rank : 68) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q812A3 | Gene names | Ankrd23, Darp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 23 (Diabetes-related ankyrin repeat protein). | |||||
|
ANK1_HUMAN
|
||||||
| NC score | 0.054350 (rank : 69) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16157, O43400, Q13768, Q59FP2, Q8N604, Q99407 | Gene names | ANK1, ANK | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin) (Ankyrin-R). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.054340 (rank : 70) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
CSKI1_MOUSE
|
||||||
| NC score | 0.054232 (rank : 71) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1031 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6P9K8, Q6ZPU2, Q8BWU2, Q8BX99, Q9CXH0 | Gene names | Caskin1, Kiaa1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
CSKI1_HUMAN
|
||||||
| NC score | 0.054091 (rank : 72) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q8WXD9, Q9P2P0 | Gene names | CASKIN1, KIAA1306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-1 (CASK-interacting protein 1). | |||||
|
ASB8_MOUSE
|
||||||
| NC score | 0.054000 (rank : 73) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT9, Q8R178 | Gene names | Asb8 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 8 (ASB-8). | |||||
|
ANR42_MOUSE
|
||||||
| NC score | 0.053995 (rank : 74) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q3V096, Q8BX55, Q8BZT3, Q8C0T6, Q8C0X5 | Gene names | Ankrd42 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 42. | |||||
|
ANKR1_MOUSE
|
||||||
| NC score | 0.053956 (rank : 75) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 316 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR42, O55014, Q3UIF7, Q3UJ39, Q792Q9 | Gene names | Ankrd1, Carp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein). | |||||
|
ANFY1_MOUSE
|
||||||
| NC score | 0.053838 (rank : 76) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q810B6, O54807, Q80TG6, Q80UH8 | Gene names | Ankfy1, Ankhzn, Kiaa1255 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and FYVE domain-containing protein 1 (Ankyrin repeats hooked to a zinc finger motif). | |||||
|
ANK1_MOUSE
|
||||||
| NC score | 0.053663 (rank : 77) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 430 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02357, P70440, P97446, P97941, Q3TZ35, Q3UH42, Q3UHP2, Q3UYY7, Q61302, Q61303, Q78E45 | Gene names | Ank1, Ank-1 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-1 (Erythrocyte ankyrin). | |||||
|
ANR41_HUMAN
|
||||||
| NC score | 0.053580 (rank : 78) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NAG6, Q8N8J8 | Gene names | ANKRD41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 41. | |||||
|
TRPA1_MOUSE
|
||||||
| NC score | 0.053473 (rank : 79) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BLA8 | Gene names | Trpa1, Anktm1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1). | |||||
|
ANKR1_HUMAN
|
||||||
| NC score | 0.053302 (rank : 80) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15327, Q96LE7 | Gene names | ANKRD1, C193, CARP, HA1A2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 1 (Cardiac ankyrin repeat protein) (Cytokine-inducible nuclear protein) (C-193). | |||||
|
ANR28_MOUSE
|
||||||
| NC score | 0.053260 (rank : 81) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q505D1 | Gene names | Ankrd28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR28_HUMAN
|
||||||
| NC score | 0.053022 (rank : 82) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 314 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15084, Q6ULS0, Q6ZT57 | Gene names | ANKRD28, KIAA0379 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 28. | |||||
|
ANR44_HUMAN
|
||||||
| NC score | 0.052894 (rank : 83) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N8A2, Q6P480, Q86VL5, Q8IZ72, Q9UFA4 | Gene names | ANKRD44 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 44. | |||||
|
TRPA1_HUMAN
|
||||||
| NC score | 0.052362 (rank : 84) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75762 | Gene names | TRPA1, ANKTM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transient receptor potential cation channel subfamily A member 1 (Ankyrin-like with transmembrane domains protein 1) (Transformation sensitive-protein p120). | |||||
|
ASB5_HUMAN
|
||||||
| NC score | 0.052273 (rank : 85) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WWX0 | Gene names | ASB5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 5 (ASB-5). | |||||
|
CDN2D_MOUSE
|
||||||
| NC score | 0.052093 (rank : 86) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60773, Q60794 | Gene names | Cdkn2d | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.051927 (rank : 87) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ASB10_HUMAN
|
||||||
| NC score | 0.051466 (rank : 88) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXI3 | Gene names | ASB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 10 (ASB-10). | |||||
|
ANR25_MOUSE
|
||||||
| NC score | 0.051421 (rank : 89) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 398 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BX02, Q8BLD5, Q91Z35 | Gene names | Ankrd25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 25. | |||||
|
ABTB2_HUMAN
|
||||||
| NC score | 0.051091 (rank : 90) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N961, Q6MZW4, Q8NB44 | Gene names | ABTB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat and BTB/POZ domain-containing protein 2. | |||||
|
INVS_HUMAN
|
||||||
| NC score | 0.051081 (rank : 91) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y283, Q5W0T6, Q8IVX8, Q9BRB9, Q9Y488, Q9Y498 | Gene names | INVS, INV, NPHP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inversin (Inversion of embryo turning homolog) (Nephrocystin-2). | |||||
|
ASB14_MOUSE
|
||||||
| NC score | 0.050682 (rank : 92) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 293 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS7 | Gene names | Asb14 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 14 (ASB-14). | |||||
|
FANK1_MOUSE
|
||||||
| NC score | 0.050481 (rank : 93) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9DAM9, Q9CUA7 | Gene names | Fank1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Fibronectin type 3 and ankyrin repeat domains 1 protein (Germ cell- specific gene 1 protein) (GSG1). | |||||
|
ANR50_HUMAN
|
||||||
| NC score | 0.050464 (rank : 94) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ULJ7, Q6N064, Q6ZSE6 | Gene names | ANKRD50, KIAA1223 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 50. | |||||
|
OSBL1_HUMAN
|
||||||
| NC score | 0.050217 (rank : 95) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BXW6, Q9BZF5, Q9NW87 | Gene names | OSBPL1A, ORP1, OSBP8, OSBPL1 | |||
|
Domain Architecture |

|
|||||
| Description | Oxysterol-binding protein-related protein 1 (OSBP-related protein 1) (ORP-1). | |||||
|
CDN2D_HUMAN
|
||||||
| NC score | 0.050193 (rank : 96) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55273, Q13102, Q6FGE9 | Gene names | CDKN2D | |||
|
Domain Architecture |

|
|||||
| Description | Cyclin-dependent kinase 4 inhibitor D (p19-INK4d). | |||||
|
ASB15_MOUSE
|
||||||
| NC score | 0.050025 (rank : 97) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
GNAS3_HUMAN
|
||||||
| NC score | 0.040487 (rank : 98) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95467, O95417 | Gene names | GNAS, GNAS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.037626 (rank : 99) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
IL7RA_MOUSE
|
||||||
| NC score | 0.036780 (rank : 100) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P16872 | Gene names | Il7r | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-7 receptor alpha chain precursor (IL-7R-alpha) (CD127 antigen). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.033422 (rank : 101) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RAI2_MOUSE
|
||||||
| NC score | 0.032379 (rank : 102) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QVY8 | Gene names | Rai2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 2 (3f8). | |||||
|
GAK6_HUMAN
|
||||||
| NC score | 0.029117 (rank : 103) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P62685 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_8p23.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K115 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK12_HUMAN
|
||||||
| NC score | 0.028836 (rank : 104) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63130, Q9UKI0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_1q22 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK2_HUMAN
|
||||||
| NC score | 0.028659 (rank : 105) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q7LDI9, Q9UKH5, Q9Y6I1, Q9YNA6, Q9YNB0 | Gene names | ERVK6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_7p22.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(HML-2.HOM) Gag protein) (HERV-K108 Gag protein) (HERV-K(C7) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK4_HUMAN
|
||||||
| NC score | 0.028555 (rank : 106) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63126, Q9UKH4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_6q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K109 Gag protein) (HERV-K(C6) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK11_HUMAN
|
||||||
| NC score | 0.028549 (rank : 107) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P63145, Q9UKI1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_22q11.21 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K101 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.028135 (rank : 108) | θ value | 0.47712 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
GAK1_HUMAN
|
||||||
| NC score | 0.027786 (rank : 109) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 268 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62683 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_12q14.1 provirus ancestral Gag polyprotein (Gag polyprotein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK5_HUMAN
|
||||||
| NC score | 0.027485 (rank : 110) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 429 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P62684 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19p13.11 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K113 Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
GAK3_HUMAN
|
||||||
| NC score | 0.027087 (rank : 111) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9YNA8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HERV-K_19q12 provirus ancestral Gag polyprotein (Gag polyprotein) (HERV-K(C19) Gag protein) [Contains: Matrix protein; Capsid protein; Nucleocapsid protein]. | |||||
|
YLPM1_HUMAN
|
||||||
| NC score | 0.026641 (rank : 112) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P49750, P49752, Q96I64, Q9P1V7 | Gene names | YLPM1, C14orf170, ZAP3 | |||
|
Domain Architecture |

|
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3) (ZAP113). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.026445 (rank : 113) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
BL1S3_HUMAN
|
||||||
| NC score | 0.025110 (rank : 114) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6QNY0 | Gene names | BLOC1S3, BLOS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Biogenesis of lysosome-related organelles complex-1 subunit 3 (BLOC subunit 3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.023196 (rank : 115) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.022213 (rank : 116) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MLL2_HUMAN
|
||||||
| NC score | 0.020680 (rank : 117) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1788 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O14686, O14687 | Gene names | MLL2, ALR | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 2 (ALL1-related protein). | |||||
|
TAB3_MOUSE
|
||||||
| NC score | 0.020308 (rank : 118) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q571K4 | Gene names | Map3k7ip3, Kiaa4135, Tab3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 3 (TAK1-binding protein 3). | |||||
|
K1958_MOUSE
|
||||||
| NC score | 0.018496 (rank : 119) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8C4P0, Q8BN62, Q8K1X8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1958 homolog. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.018208 (rank : 120) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.016876 (rank : 121) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
LPP_HUMAN
|
||||||
| NC score | 0.016085 (rank : 122) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 775 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q93052, Q8NFX5 | Gene names | LPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lipoma-preferred partner (LIM domain-containing preferred translocation partner in lipoma). | |||||
|
CASK_HUMAN
|
||||||
| NC score | 0.016064 (rank : 123) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07498, Q13575 | Gene names | CSN3, CASK, CSN10, CSNK | |||
|
Domain Architecture |

|
|||||
| Description | Kappa-casein precursor. | |||||
|
IPMK_HUMAN
|
||||||
| NC score | 0.015462 (rank : 124) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NFU5 | Gene names | IPMK, IMPK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol polyphosphate multikinase (EC 2.7.1.151) (Inositol 1,3,4,6- tetrakisphosphate 5-kinase). | |||||
|
SELPL_MOUSE
|
||||||
| NC score | 0.014343 (rank : 125) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62170 | Gene names | Selplg, Selp1, Selpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P-selectin glycoprotein ligand 1 precursor (PSGL-1) (Selectin P ligand). | |||||
|
C8AP2_MOUSE
|
||||||
| NC score | 0.014095 (rank : 126) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 405 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9WUF3, Q9CSW2 | Gene names | Casp8ap2, Flash | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
PARC_HUMAN
|
||||||
| NC score | 0.013195 (rank : 127) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWT3, O75188, Q68CP2, Q68D92, Q8N3W9, Q9BU56 | Gene names | PARC, H7AP1, KIAA0708 | |||
|
Domain Architecture |

|
|||||
| Description | p53-associated parkin-like cytoplasmic protein (UbcH7-associated protein 1). | |||||
|
NKX23_HUMAN
|
||||||
| NC score | 0.012212 (rank : 128) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TAU0, Q9NYS6 | Gene names | NKX2-3, NKX23, NKX2C | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.3 (Homeobox protein NKX-2.3) (Homeobox protein NK-2 homolog C). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.010287 (rank : 129) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.010069 (rank : 130) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.009984 (rank : 131) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
TDG_MOUSE
|
||||||
| NC score | 0.009930 (rank : 132) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56581 | Gene names | Tdg | |||
|
Domain Architecture |

|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-) (C-JUN leucine zipper interactive protein JZA-3). | |||||
|
AAKG2_MOUSE
|
||||||
| NC score | 0.009207 (rank : 133) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WG5, Q6V7V5 | Gene names | Prkag2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 5'-AMP-activated protein kinase subunit gamma-2 (AMPK gamma-2 chain) (AMPK gamma2). | |||||
|
RNF37_MOUSE
|
||||||
| NC score | 0.008516 (rank : 134) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q925F4 | Gene names | Ubox5, Rnf37, Ubce7ip5, Uip5 | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 37 (Ubiquitin-conjugating enzyme 7-interacting protein 5) (UbcM4-interacting protein 5) (U-box domain-containing protein 5). | |||||
|
NKX24_HUMAN
|
||||||
| NC score | 0.008120 (rank : 135) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H2Z4 | Gene names | NKX2-4, NKX2D | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.4 (Homeobox protein NKX2.4) (Homeobox protein NK-2 homolog D). | |||||
|
PTN23_HUMAN
|
||||||
| NC score | 0.007840 (rank : 136) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 646 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H3S7, Q7KZF8, Q8N6Z5, Q9BSR5, Q9P257, Q9UG03, Q9UMZ4 | Gene names | PTPN23, KIAA1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48) (His- domain-containing protein tyrosine phosphatase) (HD-PTP) (Protein tyrosine phosphatase TD14) (PTP-TD14). | |||||
|
PPCKC_MOUSE
|
||||||
| NC score | 0.006463 (rank : 137) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z2V4, Q3UEH3 | Gene names | Pck1, Pepck | |||
|
Domain Architecture |

|
|||||
| Description | Phosphoenolpyruvate carboxykinase, cytosolic [GTP] (EC 4.1.1.32) (Phosphoenolpyruvate carboxylase) (PEPCK-C). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.006365 (rank : 138) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.005673 (rank : 139) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
PAR3L_HUMAN
|
||||||
| NC score | 0.004571 (rank : 140) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 297 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8TEW8, Q8IUC7, Q8IUC9, Q96DK9, Q96N09, Q96NX6, Q96NX7, Q96Q29 | Gene names | PARD3B, ALS2CR19, PAR3B, PAR3L | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog B (PAR3-beta) (Partitioning-defective 3-like protein) (PAR3-L protein) (Amyotrophic lateral sclerosis 2 chromosome region candidate gene 19 protein). | |||||
|
WEE1B_HUMAN
|
||||||
| NC score | 0.003015 (rank : 141) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 836 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P0C1S8 | Gene names | WEE1B, WEE2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wee1-like protein kinase 1B (EC 2.7.10.2) (Wee1B kinase). | |||||
|
KLF17_HUMAN
|
||||||
| NC score | 0.001994 (rank : 142) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 882 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5JT82, Q86VQ7, Q8N805 | Gene names | KLF17, ZNF393 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 17 (Zinc finger protein 393). | |||||
|
KTNB1_MOUSE
|
||||||
| NC score | 0.001850 (rank : 143) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BG40, Q8CD18, Q8R1J0, Q9CWV2 | Gene names | Katnb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Katanin p80 WD40-containing subunit B1 (Katanin p80 subunit B1) (p80 katanin). | |||||
|
ACRO_HUMAN
|
||||||
| NC score | 0.001389 (rank : 144) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
LTBP4_MOUSE
|
||||||
| NC score | 0.001267 (rank : 145) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K4G1, Q8K4G0 | Gene names | Ltbp4 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 4 precursor (LTBP-4). | |||||