Please be patient as the page loads
|
DTX1_MOUSE
|
||||||
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
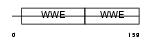
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DTX1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.987411 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86Y01, O60630, Q9BS04 | Gene names | DTX1 | |||
|
Domain Architecture |
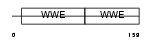
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (hDTX1). | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
DTX2_HUMAN
|
||||||
| θ value | 1.12714e-143 (rank : 3) | NC score | 0.947690 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |
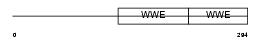
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
DTX2_MOUSE
|
||||||
| θ value | 1.57644e-137 (rank : 4) | NC score | 0.946894 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |
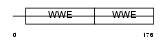
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
DTX3L_HUMAN
|
||||||
| θ value | 1.20211e-36 (rank : 5) | NC score | 0.712098 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDB6 | Gene names | DTX3L, BBAP | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2). | |||||
|
DTX3_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 6) | NC score | 0.668176 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N9I9, Q8NAU6, Q8NDS8 | Gene names | DTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3). | |||||
|
DTX3_MOUSE
|
||||||
| θ value | 3.87602e-27 (rank : 7) | NC score | 0.664997 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80V91, Q9ER06 | Gene names | Dtx3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3) (mDTX3). | |||||
|
CEL_HUMAN
|
||||||
| θ value | 0.00134147 (rank : 8) | NC score | 0.052821 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
MAGE1_HUMAN
|
||||||
| θ value | 0.00509761 (rank : 9) | NC score | 0.056811 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 0.0113563 (rank : 10) | NC score | 0.054728 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
RNF32_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 11) | NC score | 0.173437 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
ZN142_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 12) | NC score | 0.011481 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
RNF32_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 13) | NC score | 0.140080 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 14) | NC score | 0.064178 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
TS13_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.072313 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.072128 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.057491 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 18) | NC score | 0.075470 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 19) | NC score | 0.042315 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 20) | NC score | 0.050019 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | 0.125558 (rank : 21) | NC score | 0.062724 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 0.163984 (rank : 22) | NC score | 0.034603 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 0.163984 (rank : 23) | NC score | 0.061136 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 24) | NC score | 0.089878 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 25) | NC score | 0.046440 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.045612 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TNR8_HUMAN
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.036112 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |
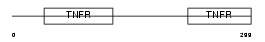
|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
FOG1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 28) | NC score | 0.024072 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
NRX2A_HUMAN
|
||||||
| θ value | 0.47712 (rank : 29) | NC score | 0.032839 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
NRX2B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 30) | NC score | 0.047371 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
RN126_MOUSE
|
||||||
| θ value | 0.47712 (rank : 31) | NC score | 0.066998 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
CBP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 32) | NC score | 0.050728 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 33) | NC score | 0.034611 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 34) | NC score | 0.043892 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.064953 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
RN126_HUMAN
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.067619 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
MAGD1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.023383 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.039820 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RN165_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.079577 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
CTDP1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.036094 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
ERRFI_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.029747 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
MK04_MOUSE
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.001171 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
SEM6C_MOUSE
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.019505 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
TCGAP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 44) | NC score | 0.024536 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
TF65_HUMAN
|
||||||
| θ value | 1.38821 (rank : 45) | NC score | 0.026598 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 46) | NC score | 0.039027 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.047638 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
ZN516_MOUSE
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.011320 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.010378 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.071054 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.058904 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
FOXL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.012148 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |
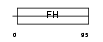
|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.043252 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
CS007_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.040241 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
GR65_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.023462 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.055159 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
RGS12_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.023875 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.038376 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
STAR8_HUMAN
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.014583 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
ZN467_MOUSE
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.002025 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8JZL0, Q9JJ98 | Gene names | Znf467, Ezi, Zfp467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467 (Endothelial cell-derived zinc finger protein) (EZI). | |||||
|
ZN646_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.005384 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
ZSWM2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.032421 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
CABIN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.058980 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
KIFC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 64) | NC score | 0.005496 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BW19, O60887, Q14834, Q9UQP7 | Gene names | KIFC1, HSET, KNSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC1 (Kinesin-like protein 2) (Kinesin-related protein HSET). | |||||
|
RN125_MOUSE
|
||||||
| θ value | 4.03905 (rank : 65) | NC score | 0.040310 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 4.03905 (rank : 66) | NC score | 0.020417 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TRI30_MOUSE
|
||||||
| θ value | 4.03905 (rank : 67) | NC score | 0.026948 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15533, Q99K55, Q99PQ7, Q99PQ8, Q99PQ9 | Gene names | Trim30, Rpt-1, Rpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 30 (Down regulatory protein of interleukin 2 receptor). | |||||
|
ZEP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.012269 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
CSKI2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 69) | NC score | 0.021451 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
EFS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 70) | NC score | 0.027288 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
JIP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 71) | NC score | 0.018589 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
KCNQ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 72) | NC score | 0.012900 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
KSYK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 73) | NC score | -0.001188 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||
|
PDLI5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 74) | NC score | 0.011147 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
PTN23_MOUSE
|
||||||
| θ value | 5.27518 (rank : 75) | NC score | 0.029182 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
RN175_HUMAN
|
||||||
| θ value | 5.27518 (rank : 76) | NC score | 0.038520 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N4F7 | Gene names | RNF175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 175. | |||||
|
HD_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.027297 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
HD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.025599 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 6.88961 (rank : 79) | NC score | 0.040664 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 80) | NC score | 0.016673 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
PI5PA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 81) | NC score | 0.033939 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 82) | NC score | 0.023923 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
PRRT3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 83) | NC score | 0.035819 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
ROBO4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 84) | NC score | 0.009084 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |
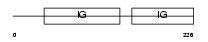
|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
SPF45_MOUSE
|
||||||
| θ value | 6.88961 (rank : 85) | NC score | 0.017721 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
TF65_MOUSE
|
||||||
| θ value | 6.88961 (rank : 86) | NC score | 0.016876 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
ZMY11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 87) | NC score | 0.019033 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
ZN133_HUMAN
|
||||||
| θ value | 6.88961 (rank : 88) | NC score | 0.001433 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52736, Q9BUV2, Q9H443 | Gene names | ZNF133 | |||
|
Domain Architecture |
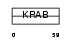
|
|||||
| Description | Zinc finger protein 133. | |||||
|
BIN1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 89) | NC score | 0.023871 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 8.99809 (rank : 90) | NC score | 0.007866 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 91) | NC score | 0.002822 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
EP400_MOUSE
|
||||||
| θ value | 8.99809 (rank : 92) | NC score | 0.026201 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
IER5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 93) | NC score | 0.027213 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
IF4G1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 94) | NC score | 0.021402 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 95) | NC score | 0.023001 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MMP11_MOUSE
|
||||||
| θ value | 8.99809 (rank : 96) | NC score | 0.004995 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
TM108_MOUSE
|
||||||
| θ value | 8.99809 (rank : 97) | NC score | 0.041596 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
ZC3H6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 98) | NC score | 0.023003 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
ZN509_MOUSE
|
||||||
| θ value | 8.99809 (rank : 99) | NC score | 0.003080 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.058137 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CIC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.052929 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CN032_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.068324 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
CN032_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.072037 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.051121 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.060870 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
GGN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.054824 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.059147 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
MBD6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.061894 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
MUC7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.070861 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
PELP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.055262 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.070994 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.067063 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.052326 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.074572 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.084515 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.050253 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.065042 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
PRR13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.064261 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
RING1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.051835 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |
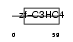
|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.050298 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.063118 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.061211 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SMR1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.053171 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
SMR3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.059521 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
TAF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.062752 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.054812 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.061433 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
WASIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.064987 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
WASIP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.064365 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
WASL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.052694 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.059564 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.059613 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WIRE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.057544 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
WIRE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.056218 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050715 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
ZN207_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.054645 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
DTX1_HUMAN
|
||||||
| NC score | 0.987411 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q86Y01, O60630, Q9BS04 | Gene names | DTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (hDTX1). | |||||
|
DTX2_HUMAN
|
||||||
| NC score | 0.947690 (rank : 3) | θ value | 1.12714e-143 (rank : 3) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
DTX2_MOUSE
|
||||||
| NC score | 0.946894 (rank : 4) | θ value | 1.57644e-137 (rank : 4) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
DTX3L_HUMAN
|
||||||
| NC score | 0.712098 (rank : 5) | θ value | 1.20211e-36 (rank : 5) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TDB6 | Gene names | DTX3L, BBAP | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2). | |||||
|
DTX3_HUMAN
|
||||||
| NC score | 0.668176 (rank : 6) | θ value | 3.87602e-27 (rank : 6) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8N9I9, Q8NAU6, Q8NDS8 | Gene names | DTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3). | |||||
|
DTX3_MOUSE
|
||||||
| NC score | 0.664997 (rank : 7) | θ value | 3.87602e-27 (rank : 7) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80V91, Q9ER06 | Gene names | Dtx3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3) (mDTX3). | |||||
|
RNF32_HUMAN
|
||||||
| NC score | 0.173437 (rank : 8) | θ value | 0.0148317 (rank : 11) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
RNF32_MOUSE
|
||||||
| NC score | 0.140080 (rank : 9) | θ value | 0.0252991 (rank : 13) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.089878 (rank : 10) | θ value | 0.21417 (rank : 24) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.084515 (rank : 11) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
RN165_HUMAN
|
||||||
| NC score | 0.079577 (rank : 12) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.075470 (rank : 13) | θ value | 0.0961366 (rank : 18) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.074572 (rank : 14) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
TS13_MOUSE
|
||||||
| NC score | 0.072313 (rank : 15) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01755 | Gene names | Tcp11 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protein PBS13. | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.072128 (rank : 16) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
CN032_MOUSE
|
||||||
| NC score | 0.072037 (rank : 17) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BH93, Q3TF12, Q3TGL0, Q8CC90 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32 homolog. | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.071054 (rank : 18) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.070994 (rank : 19) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
MUC7_HUMAN
|
||||||
| NC score | 0.070861 (rank : 20) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8TAX7 | Gene names | MUC7, MG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mucin-7 precursor (MUC-7) (Salivary mucin-7) (Apo-MG2). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.068324 (rank : 21) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
RN126_HUMAN
|
||||||
| NC score | 0.067619 (rank : 22) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BV68, Q9NWX1 | Gene names | RNF126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.067063 (rank : 23) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
RN126_MOUSE
|
||||||
| NC score | 0.066998 (rank : 24) | θ value | 0.47712 (rank : 31) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q91YL2 | Gene names | Rnf126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 126. | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.065042 (rank : 25) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
WASIP_HUMAN
|
||||||
| NC score | 0.064987 (rank : 26) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | O43516, Q15220, Q6MZU9, Q9BU37, Q9UNP1 | Gene names | WASPIP, WIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) (PRPL-2 protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.064953 (rank : 27) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
WASIP_MOUSE
|
||||||
| NC score | 0.064365 (rank : 28) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 392 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8K1I7, Q3U0U8 | Gene names | Waspip, Wip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein). | |||||
|
PRR13_HUMAN
|
||||||
| NC score | 0.064261 (rank : 29) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9NZ81, Q6FIG7, Q6MZP8, Q6NXQ6, Q6PKF9 | Gene names | PRR13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 13. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.064178 (rank : 30) | θ value | 0.0431538 (rank : 14) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.063118 (rank : 31) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
TAF4_HUMAN
|
||||||
| NC score | 0.062752 (rank : 32) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | O00268, Q5TBP6, Q99721, Q9BR40, Q9BX42 | Gene names | TAF4, TAF2C, TAF2C1, TAF4A, TAFII130, TAFII135 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 4 (TBP-associated factor 4) (Transcription initiation factor TFIID 135 kDa subunit) (TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130) (TAFII130). | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.062724 (rank : 33) | θ value | 0.125558 (rank : 21) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MBD6_HUMAN
|
||||||
| NC score | 0.061894 (rank : 34) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q96DN6, Q8N3M0, Q8NA81, Q96Q00 | Gene names | MBD6, KIAA1887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 6 (Methyl-CpG-binding protein MBD6). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.061433 (rank : 35) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.061211 (rank : 36) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.061136 (rank : 37) | θ value | 0.163984 (rank : 23) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.060870 (rank : 38) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.059613 (rank : 39) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.059564 (rank : 40) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
SMR3A_HUMAN
|
||||||
| NC score | 0.059521 (rank : 41) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99954 | Gene names | SMR3A, PBI, PROL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 3 homolog A precursor (Proline-rich protein 5) (Proline-rich protein PBI). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.059147 (rank : 42) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
CABIN_HUMAN
|
||||||
| NC score | 0.058980 (rank : 43) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 490 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9Y6J0, Q9Y460 | Gene names | CABIN1, KIAA0330 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin-binding protein Cabin 1 (Calcineurin inhibitor) (CAIN). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.058904 (rank : 44) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.058137 (rank : 45) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
WIRE_HUMAN
|
||||||
| NC score | 0.057544 (rank : 46) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8TF74, Q658J8, Q71RE1, Q8TE44 | Gene names | WIRE, WICH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein) (WIP- and CR16-homologous protein). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.057491 (rank : 47) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
MAGE1_HUMAN
|
||||||
| NC score | 0.056811 (rank : 48) | θ value | 0.00509761 (rank : 9) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1389 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9HCI5, Q86TG0, Q8TD92, Q9H216 | Gene names | MAGEE1, HCA1, KIAA1587 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanoma-associated antigen E1 (MAGE-E1 antigen) (Hepatocellular carcinoma-associated protein 1). | |||||
|
WIRE_MOUSE
|
||||||
| NC score | 0.056218 (rank : 49) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q6PEV3 | Gene names | Wire | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WIP-related protein (WASP-interacting protein-related protein). | |||||
|
PELP1_HUMAN
|
||||||
| NC score | 0.055262 (rank : 50) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8IZL8, O15450, Q5EGN3, Q6NTE6, Q96FT1, Q9BU60 | Gene names | PELP1, HMX3, MNAR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor) (Transcription factor HMX3). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.055159 (rank : 51) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.054824 (rank : 52) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.054812 (rank : 53) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.054728 (rank : 54) | θ value | 0.0113563 (rank : 10) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.054645 (rank : 55) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
SMR1_MOUSE
|
||||||
| NC score | 0.053171 (rank : 56) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61900 | Gene names | Smr1, Msg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Submaxillary gland androgen-regulated protein 1 precursor (Salivary protein MSG1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.052929 (rank : 57) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CEL_HUMAN
|
||||||
| NC score | 0.052821 (rank : 58) | θ value | 0.00134147 (rank : 8) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 896 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P19835, Q16398 | Gene names | CEL, BAL | |||
|
Domain Architecture |

|
|||||
| Description | Bile salt-activated lipase precursor (EC 3.1.1.3) (EC 3.1.1.13) (BAL) (Bile salt-stimulated lipase) (BSSL) (Carboxyl ester lipase) (Sterol esterase) (Cholesterol esterase) (Pancreatic lysophospholipase). | |||||
|
WASL_MOUSE
|
||||||
| NC score | 0.052694 (rank : 59) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q91YD9 | Gene names | Wasl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neural Wiskott-Aldrich syndrome protein (N-WASP). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.052326 (rank : 60) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RING1_HUMAN
|
||||||
| NC score | 0.051835 (rank : 61) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.051121 (rank : 62) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
CBP_MOUSE
|
||||||
| NC score | 0.050728 (rank : 63) | θ value | 0.62314 (rank : 32) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1051 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P45481 | Gene names | Crebbp, Cbp | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.050715 (rank : 64) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.050298 (rank : 65) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.050253 (rank : 66) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.050019 (rank : 67) | θ value | 0.0961366 (rank : 20) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.047638 (rank : 68) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
NRX2B_HUMAN
|
||||||
| NC score | 0.047371 (rank : 69) | θ value | 0.47712 (rank : 30) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P58401 | Gene names | NRXN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurexin-2-beta precursor (Neurexin II-beta). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.046440 (rank : 70) | θ value | 0.279714 (rank : 25) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.045612 (rank : 71) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.043892 (rank : 72) | θ value | 0.62314 (rank : 34) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.043252 (rank : 73) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.042315 (rank : 74) | θ value | 0.0961366 (rank : 19) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
TM108_MOUSE
|
||||||
| NC score | 0.041596 (rank : 75) | θ value | 8.99809 (rank : 97) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q8BHE4, Q80WR9 | Gene names | Tmem108 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protein 108 precursor. | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.040664 (rank : 76) | θ value | 6.88961 (rank : 79) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
RN125_MOUSE
|
||||||
| NC score | 0.040310 (rank : 77) | θ value | 4.03905 (rank : 65) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D9R0, Q52KL4, Q8C7F2 | Gene names | Rnf125 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 125 (EC 6.3.2.-). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.040241 (rank : 78) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.039820 (rank : 79) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.039027 (rank : 80) | θ value | 1.38821 (rank : 46) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
RN175_HUMAN
|
||||||
| NC score | 0.038520 (rank : 81) | θ value | 5.27518 (rank : 76) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8N4F7 | Gene names | RNF175 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 175. | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.038376 (rank : 82) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TNR8_HUMAN
|
||||||
| NC score | 0.036112 (rank : 83) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28908 | Gene names | TNFRSF8, CD30 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 8 precursor (CD30L receptor) (Lymphocyte activation antigen CD30) (KI-1 antigen). | |||||
|
CTDP1_HUMAN
|
||||||
| NC score | 0.036094 (rank : 84) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Y5B0, Q7Z644, Q96BZ1, Q9Y6F5 | Gene names | CTDP1, FCP1 | |||
|
Domain Architecture |

|
|||||
| Description | RNA polymerase II subunit A C-terminal domain phosphatase (EC 3.1.3.16) (TFIIF-associating CTD phosphatase). | |||||
|
PRRT3_HUMAN
|
||||||
| NC score | 0.035819 (rank : 85) | θ value | 6.88961 (rank : 83) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q5FWE3, Q49AD0, Q6UXY6, Q8NBC9 | Gene names | PRRT3, UNQ5823/PRO19642 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich transmembrane protein 3. | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.034611 (rank : 86) | θ value | 0.62314 (rank : 33) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.034603 (rank : 87) | θ value | 0.163984 (rank : 22) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
PI5PA_HUMAN
|
||||||
| NC score | 0.033939 (rank : 88) | θ value | 6.88961 (rank : 81) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 749 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15735, Q32M61, Q6ZTH6, Q8N902, Q9UDT9 | Gene names | PIB5PA, PIPP | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A (EC 3.1.3.56). | |||||
|
NRX2A_HUMAN
|
||||||
| NC score | 0.032839 (rank : 89) | θ value | 0.47712 (rank : 29) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9P2S2, Q9Y2D6 | Gene names | NRXN2, KIAA0921 | |||
|
Domain Architecture |

|
|||||
| Description | Neurexin-2-alpha precursor (Neurexin II-alpha). | |||||
|
ZSWM2_HUMAN
|
||||||
| NC score | 0.032421 (rank : 90) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8NEG5 | Gene names | ZSWIM2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger SWIM domain-containing protein 2. | |||||
|
ERRFI_HUMAN
|
||||||
| NC score | 0.029747 (rank : 91) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UJM3, Q9NTG9, Q9UD05 | Gene names | ERRFI1, MIG6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERBB receptor feedback inhibitor 1 (Mitogen-inducible gene 6 protein) (Mig-6). | |||||
|
PTN23_MOUSE
|
||||||
| NC score | 0.029182 (rank : 92) | θ value | 5.27518 (rank : 75) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 726 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q6PB44, Q69ZJ0, Q8R1Z5, Q923E6 | Gene names | Ptpn23, Kiaa1471 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 23 (EC 3.1.3.48). | |||||
|
HD_HUMAN
|
||||||
| NC score | 0.027297 (rank : 93) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P42858, Q9UQB7 | Gene names | HD, IT15 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein) (HD protein). | |||||
|
EFS_MOUSE
|
||||||
| NC score | 0.027288 (rank : 94) | θ value | 5.27518 (rank : 70) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q64355 | Gene names | Efs, Sin | |||
|
Domain Architecture |

|
|||||
| Description | Embryonal Fyn-associated substrate (SRC-interacting protein) (Signal- integrating protein). | |||||
|
IER5_MOUSE
|
||||||
| NC score | 0.027213 (rank : 95) | θ value | 8.99809 (rank : 93) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O89113, Q8CGI0 | Gene names | Ier5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 5 protein. | |||||
|
TRI30_MOUSE
|
||||||
| NC score | 0.026948 (rank : 96) | θ value | 4.03905 (rank : 67) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P15533, Q99K55, Q99PQ7, Q99PQ8, Q99PQ9 | Gene names | Trim30, Rpt-1, Rpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 30 (Down regulatory protein of interleukin 2 receptor). | |||||
|
TF65_HUMAN
|
||||||
| NC score | 0.026598 (rank : 97) | θ value | 1.38821 (rank : 45) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q04206, Q6SLK1 | Gene names | RELA, NFKB3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
EP400_MOUSE
|
||||||
| NC score | 0.026201 (rank : 98) | θ value | 8.99809 (rank : 92) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8CHI8, Q80TC8, Q8BXI5, Q8BYW3, Q8C0P6, Q8CHI7, Q8VDF4, Q9DA54 | Gene names | Ep400, Kiaa1498 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E1A-binding protein p400 (EC 3.6.1.-) (p400 kDa SWI2/SNF2-related protein) (Domino homolog) (mDomino). | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.025599 (rank : 99) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
TCGAP_HUMAN
|
||||||
| NC score | 0.024536 (rank : 100) | θ value | 1.38821 (rank : 44) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 872 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O14559, O14552, O14560, Q6ZSP6, Q96CP3, Q9NT23 | Gene names | SNX26, TCGAP | |||
|
Domain Architecture |

|
|||||
| Description | TC10/CDC42 GTPase-activating protein (Sorting nexin-26). | |||||
|
FOG1_MOUSE
|
||||||
| NC score | 0.024072 (rank : 101) | θ value | 0.47712 (rank : 28) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1237 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O35615 | Gene names | Zfpm1, Fog, Fog1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.023923 (rank : 102) | θ value | 6.88961 (rank : 82) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RGS12_HUMAN
|
||||||
| NC score | 0.023875 (rank : 103) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O14924, O14922, O14923, O43510, O75338 | Gene names | RGS12 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 12 (RGS12). | |||||
|
BIN1_HUMAN
|
||||||
| NC score | 0.023871 (rank : 104) | θ value | 8.99809 (rank : 89) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O00499, O00297, O00545, O43867, O60552, O60553, O60554, O60555, O75514, O75515, O75516, O75517, O75518, Q92944, Q99688 | Gene names | BIN1, AMPHL | |||
|
Domain Architecture |

|
|||||
| Description | Myc box-dependent-interacting protein 1 (Bridging integrator 1) (Amphiphysin-like protein) (Amphiphysin II) (Box-dependent myc- interacting protein 1). | |||||
|
GR65_HUMAN
|
||||||
| NC score | 0.023462 (rank : 105) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9BQQ3, Q8N272, Q96H42 | Gene names | GORASP1, GRASP65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 1 (Golgi reassembly-stacking protein of 65 kDa) (GRASP65) (Golgi peripheral membrane protein p65) (Golgi phosphoprotein 5) (GOLPH5). | |||||
|
MAGD1_HUMAN
|
||||||
| NC score | 0.023383 (rank : 106) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y5V3, Q5VSH6, Q8IZ84, Q8WY92, Q9H352, Q9HBT4, Q9UF36 | Gene names | MAGED1, NRAGE | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D1 (MAGE-D1 antigen) (Neurotrophin receptor-interacting MAGE homolog) (MAGE tumor antigen CCF). | |||||
|
ZC3H6_HUMAN
|
||||||
| NC score | 0.023003 (rank : 107) | θ value | 8.99809 (rank : 98) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P61129 | Gene names | ZC3H6, KIAA2035, ZC3HDC6 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH domain-containing protein 6. | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.023001 (rank : 108) | θ value | 8.99809 (rank : 95) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
CSKI2_MOUSE
|
||||||
| NC score | 0.021451 (rank : 109) | θ value | 5.27518 (rank : 69) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 812 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8VHK1, Q6ZPX1, Q7TS77 | Gene names | Caskin2, Kiaa1139 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Caskin-2. | |||||
|
IF4G1_MOUSE
|
||||||
| NC score | 0.021402 (rank : 110) | θ value | 8.99809 (rank : 94) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q6NZJ6, Q6NZN8, Q8BW99 | Gene names | Eif4g1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.020417 (rank : 111) | θ value | 4.03905 (rank : 66) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
SEM6C_MOUSE
|
||||||
| NC score | 0.019505 (rank : 112) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9WTM3 | Gene names | Sema6c, Semay | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-6C precursor (Semaphorin Y) (Sema Y). | |||||
|
ZMY11_HUMAN
|
||||||
| NC score | 0.019033 (rank : 113) | θ value | 6.88961 (rank : 87) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15326 | Gene names | ZMYND11, BS69 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger MYND domain-containing protein 11 (Adenovirus 5 E1A- binding protein) (BS69 protein). | |||||
|
JIP2_HUMAN
|
||||||
| NC score | 0.018589 (rank : 114) | θ value | 5.27518 (rank : 71) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q13387, Q96G62, Q99771, Q9NZ59, Q9UKQ4 | Gene names | MAPK8IP2, IB2, JIP2 | |||
|
Domain Architecture |

|
|||||
| Description | C-jun-amino-terminal kinase-interacting protein 2 (JNK-interacting protein 2) (JIP-2) (JNK MAP kinase scaffold protein 2) (Islet-brain-2) (IB-2) (Mitogen-activated protein kinase 8-interacting protein 2). | |||||
|
SPF45_MOUSE
|
||||||
| NC score | 0.017721 (rank : 115) | θ value | 6.88961 (rank : 85) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8JZX4, O75939 | Gene names | Rbm17, Spf45 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 45 (45 kDa-splicing factor) (RNA-binding motif protein 17). | |||||
|
TF65_MOUSE
|
||||||
| NC score | 0.016876 (rank : 116) | θ value | 6.88961 (rank : 86) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q04207, Q62025 | Gene names | Rela, Nfkb3 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor p65 (Nuclear factor NF-kappa-B p65 subunit). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.016673 (rank : 117) | θ value | 6.88961 (rank : 80) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
STAR8_HUMAN
|
||||||
| NC score | 0.014583 (rank : 118) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q92502, Q5JST0 | Gene names | STARD8, KIAA0189 | |||
|
Domain Architecture |

|
|||||
| Description | StAR-related lipid transfer protein 8 (StARD8) (START domain- containing protein 8). | |||||
|
KCNQ1_MOUSE
|
||||||
| NC score | 0.012900 (rank : 119) | θ value | 5.27518 (rank : 72) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P97414, O88702, Q7TNZ1, Q7TPL7, Q80VR7 | Gene names | Kcnq1, Kcna9, Kvlqt1 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium voltage-gated channel subfamily KQT member 1 (Voltage-gated potassium channel subunit Kv7.1) (IKs producing slow voltage-gated potassium channel subunit alpha KvLQT1) (KQT-like 1). | |||||
|
ZEP1_HUMAN
|
||||||
| NC score | 0.012269 (rank : 120) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1036 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P15822, Q14122 | Gene names | HIVEP1, ZNF40 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 40 (Human immunodeficiency virus type I enhancer- binding protein 1) (HIV-EP1) (Major histocompatibility complex-binding protein 1) (MBP-1) (Positive regulatory domain II-binding factor 1) (PRDII-BF1). | |||||
|
FOXL1_MOUSE
|
||||||
| NC score | 0.012148 (rank : 121) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q64731 | Gene names | Foxl1, Fkh6, Fkhl11 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L1 (Forkhead-related protein FKHL11) (Transcription factor FKH-6). | |||||
|
ZN142_HUMAN
|
||||||
| NC score | 0.011481 (rank : 122) | θ value | 0.0193708 (rank : 12) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P52746, Q92510 | Gene names | ZNF142, KIAA0236 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 142 (HA4654). | |||||
|
ZN516_MOUSE
|
||||||
| NC score | 0.011320 (rank : 123) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q7TSH3 | Gene names | Znf516, Kiaa0222, Zfp516 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 516. | |||||
|
PDLI5_HUMAN
|
||||||
| NC score | 0.011147 (rank : 124) | θ value | 5.27518 (rank : 74) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 322 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96HC4, O60705 | Gene names | PDLIM5, ENH | |||
|
Domain Architecture |

|
|||||
| Description | PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein). | |||||
|
ATS14_HUMAN
|
||||||
| NC score | 0.010378 (rank : 125) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
ROBO4_MOUSE
|
||||||
| NC score | 0.009084 (rank : 126) | θ value | 6.88961 (rank : 84) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8C310, Q9DBW1 | Gene names | Robo4 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 4 precursor. | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.007866 (rank : 127) | θ value | 8.99809 (rank : 90) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
KIFC1_HUMAN
|
||||||
| NC score | 0.005496 (rank : 128) | θ value | 4.03905 (rank : 64) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 545 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BW19, O60887, Q14834, Q9UQP7 | Gene names | KIFC1, HSET, KNSL2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIFC1 (Kinesin-like protein 2) (Kinesin-related protein HSET). | |||||
|
ZN646_HUMAN
|
||||||
| NC score | 0.005384 (rank : 129) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 813 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15015 | Gene names | ZNF646, KIAA0296 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 646. | |||||
|
MMP11_MOUSE
|
||||||
| NC score | 0.004995 (rank : 130) | θ value | 8.99809 (rank : 96) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02853 | Gene names | Mmp11 | |||
|
Domain Architecture |

|
|||||
| Description | Stromelysin-3 precursor (EC 3.4.24.-) (ST3) (SL-3) (Matrix metalloproteinase-11) (MMP-11). | |||||
|
ZN509_MOUSE
|
||||||
| NC score | 0.003080 (rank : 131) | θ value | 8.99809 (rank : 99) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 990 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8BXX2, Q8CID4, Q8K2Z6 | Gene names | Znf509, Zfp509 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 509. | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.002822 (rank : 132) | θ value | 8.99809 (rank : 91) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
ZN467_MOUSE
|
||||||
| NC score | 0.002025 (rank : 133) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 794 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8JZL0, Q9JJ98 | Gene names | Znf467, Ezi, Zfp467 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 467 (Endothelial cell-derived zinc finger protein) (EZI). | |||||
|
ZN133_HUMAN
|
||||||
| NC score | 0.001433 (rank : 134) | θ value | 6.88961 (rank : 88) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P52736, Q9BUV2, Q9H443 | Gene names | ZNF133 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 133. | |||||
|
MK04_MOUSE
|
||||||
| NC score | 0.001171 (rank : 135) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6P5G0 | Gene names | Mapk4, Erk4, Prkm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase 4 (EC 2.7.11.24) (Extracellular signal-regulated kinase 4) (ERK-4). | |||||
|
KSYK_MOUSE
|
||||||
| NC score | -0.001188 (rank : 136) | θ value | 5.27518 (rank : 73) | |||
| Query Neighborhood Hits | 99 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48025 | Gene names | Syk | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase SYK (EC 2.7.10.2) (Spleen tyrosine kinase). | |||||