Please be patient as the page loads
|
DTX2_MOUSE
|
||||||
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |
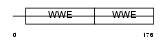
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DTX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.985233 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |
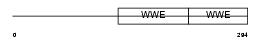
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
DTX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 1.57644e-137 (rank : 3) | NC score | 0.946894 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
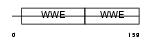
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
DTX1_HUMAN
|
||||||
| θ value | 4.58675e-137 (rank : 4) | NC score | 0.955285 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86Y01, O60630, Q9BS04 | Gene names | DTX1 | |||
|
Domain Architecture |
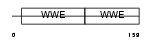
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (hDTX1). | |||||
|
DTX3L_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 5) | NC score | 0.731014 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDB6 | Gene names | DTX3L, BBAP | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2). | |||||
|
DTX3_HUMAN
|
||||||
| θ value | 5.0505e-35 (rank : 6) | NC score | 0.697445 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N9I9, Q8NAU6, Q8NDS8 | Gene names | DTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3). | |||||
|
DTX3_MOUSE
|
||||||
| θ value | 1.46948e-34 (rank : 7) | NC score | 0.694441 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V91, Q9ER06 | Gene names | Dtx3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3) (mDTX3). | |||||
|
RNF32_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 8) | NC score | 0.155558 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 9) | NC score | 0.046285 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.029431 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
HRSL5_MOUSE
|
||||||
| θ value | 0.365318 (rank : 11) | NC score | 0.032757 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPX5, Q9CVQ2, Q9CVS0 | Gene names | Hrasls5, Hrlp5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.055682 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
TBX10_HUMAN
|
||||||
| θ value | 0.47712 (rank : 13) | NC score | 0.014736 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75333, Q86XS3 | Gene names | TBX10, TBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10). | |||||
|
FOXM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 14) | NC score | 0.020022 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
MAST4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.005009 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 16) | NC score | 0.026784 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
ULK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 17) | NC score | 0.003349 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
CIP4_HUMAN
|
||||||
| θ value | 1.06291 (rank : 18) | NC score | 0.024726 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
K0460_MOUSE
|
||||||
| θ value | 1.06291 (rank : 19) | NC score | 0.027113 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
RO52_MOUSE
|
||||||
| θ value | 1.06291 (rank : 20) | NC score | 0.015222 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62191 | Gene names | Trim21, Ro52, Ssa1 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif- containing protein 21). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 1.38821 (rank : 21) | NC score | 0.032279 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
GLI1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.002510 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
NU214_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.027715 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
GORS2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.022195 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
RNF32_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.113530 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.031323 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
EHMT1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.006704 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.026655 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
NUMB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.016849 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.035325 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ZN318_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.019072 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.046746 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.046710 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
DZIP3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.033271 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
KI2L4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.008331 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |
|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.019383 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.006454 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
CSRP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.022654 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.022541 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
FOXL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.009854 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88470, Q8K3X9 | Gene names | Foxl2, Pfrk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein L2 (Pituitary forkhead factor) (P-Frk). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.015739 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.018783 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.026208 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
SMRA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.031206 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
ARNT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 45) | NC score | 0.007490 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 46) | NC score | 0.015860 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 47) | NC score | 0.017430 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 5.27518 (rank : 48) | NC score | 0.020601 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
CAC1H_MOUSE
|
||||||
| θ value | 5.27518 (rank : 49) | NC score | 0.007717 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
CRSP7_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.019390 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
CSRP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.018381 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16527, Q93030 | Gene names | CSRP2, LMO5, SMLIM | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Smooth muscle cell LIM protein) (SmLIM) (LIM-only protein 5). | |||||
|
CSRP2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.018388 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97314, Q9CZG5 | Gene names | Csrp2, Dlp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1). | |||||
|
EFNA4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.011860 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08542, O55218 | Gene names | Efna4, Epl4, Eplg4, Lerk4 | |||
|
Domain Architecture |
|
|||||
| Description | Ephrin-A4 precursor (EPH-related receptor tyrosine kinase ligand 4) (LERK-4). | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.011942 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
M4K4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.000552 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.022178 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
RAI1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.012283 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.029888 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
ZN294_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.030897 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
ZN294_MOUSE
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.030961 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6A009, Q6PIW6, Q8BZ44 | Gene names | Znf294, Kiaa0714, zfp294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 294 (Zfp-294). | |||||
|
CO4A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.011002 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P0C0L4, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q5JQM8, Q9NPK5, Q9UIP5 | Gene names | C4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-A precursor (Acidic complement C4) [Contains: Complement C4 beta chain; Complement C4-A alpha chain; C4a anaphylatoxin; C4b-A; C4d-A; Complement C4 gamma chain]. | |||||
|
CO4B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.010996 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P0C0L5, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q9NPK5, Q9UIP5 | Gene names | C4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-B precursor (Basic complement C4) [Contains: Complement C4 beta chain; Complement C4-B alpha chain; C4a anaphylatoxin; C4b-B; C4d-B; Complement C4 gamma chain]. | |||||
|
EFHD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.009712 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |
|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
FOXL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.008270 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58012, Q4ZGJ3 | Gene names | FOXL2 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein L2. | |||||
|
GAB1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009463 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
JHD2B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.009287 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
PTER_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.022321 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BW5, Q9BY46 | Gene names | PTER | |||
|
Domain Architecture |

|
|||||
| Description | Phosphotriesterase-related protein (Parathion hydrolase-related protein) (HPHRP). | |||||
|
TRI30_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.031626 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15533, Q99K55, Q99PQ7, Q99PQ8, Q99PQ9 | Gene names | Trim30, Rpt-1, Rpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 30 (Down regulatory protein of interleukin 2 receptor). | |||||
|
TTLL4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.009490 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
C1TC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.009271 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922D8 | Gene names | Mthfd1 | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
|
F100B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.010931 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYN6 | Gene names | FAM100B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100B. | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.021233 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
PKHA6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.010133 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RAPSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.023839 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
RAPSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.024275 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
ZCC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.009533 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
RING1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.056971 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.055960 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
RN165_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.056988 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.058208 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.065293 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
DTX2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
DTX2_HUMAN
|
||||||
| NC score | 0.985233 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
DTX1_HUMAN
|
||||||
| NC score | 0.955285 (rank : 3) | θ value | 4.58675e-137 (rank : 4) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86Y01, O60630, Q9BS04 | Gene names | DTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (hDTX1). | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.946894 (rank : 4) | θ value | 1.57644e-137 (rank : 3) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
DTX3L_HUMAN
|
||||||
| NC score | 0.731014 (rank : 5) | θ value | 1.57e-36 (rank : 5) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TDB6 | Gene names | DTX3L, BBAP | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2). | |||||
|
DTX3_HUMAN
|
||||||
| NC score | 0.697445 (rank : 6) | θ value | 5.0505e-35 (rank : 6) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8N9I9, Q8NAU6, Q8NDS8 | Gene names | DTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3). | |||||
|
DTX3_MOUSE
|
||||||
| NC score | 0.694441 (rank : 7) | θ value | 1.46948e-34 (rank : 7) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80V91, Q9ER06 | Gene names | Dtx3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3) (mDTX3). | |||||
|
RNF32_HUMAN
|
||||||
| NC score | 0.155558 (rank : 8) | θ value | 0.0193708 (rank : 8) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
RNF32_MOUSE
|
||||||
| NC score | 0.113530 (rank : 9) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.065293 (rank : 10) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.058208 (rank : 11) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
RN165_HUMAN
|
||||||
| NC score | 0.056988 (rank : 12) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
RING1_HUMAN
|
||||||
| NC score | 0.056971 (rank : 13) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.055960 (rank : 14) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.055682 (rank : 15) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.046746 (rank : 16) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.046710 (rank : 17) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
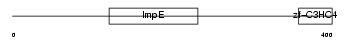
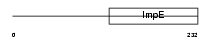
TRIPC_HUMAN
|
||||||
| NC score | 0.046285 (rank : 18) | θ value | 0.0563607 (rank : 9) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.035325 (rank : 19) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
DZIP3_HUMAN
|
||||||
| NC score | 0.033271 (rank : 20) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86Y13, O75162, Q6P3R9, Q6PH82, Q86Y14, Q86Y15, Q86Y16, Q8IWI0, Q96RS9 | Gene names | DZIP3, KIAA0675 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein DZIP3 (EC 6.3.2.-) (DAZ-interacting protein 3) (RNA-binding ubiquitin ligase of 138 kDa) (hRUL138). | |||||
|
HRSL5_MOUSE
|
||||||
| NC score | 0.032757 (rank : 21) | θ value | 0.365318 (rank : 11) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CPX5, Q9CVQ2, Q9CVS0 | Gene names | Hrasls5, Hrlp5 | |||
|
Domain Architecture |

|
|||||
| Description | HRAS-like suppressor 5 (H-rev107-like protein 5). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.032279 (rank : 22) | θ value | 1.38821 (rank : 21) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TRI30_MOUSE
|
||||||
| NC score | 0.031626 (rank : 23) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P15533, Q99K55, Q99PQ7, Q99PQ8, Q99PQ9 | Gene names | Trim30, Rpt-1, Rpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 30 (Down regulatory protein of interleukin 2 receptor). | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.031323 (rank : 24) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
SMRA3_HUMAN
|
||||||
| NC score | 0.031206 (rank : 25) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
ZN294_MOUSE
|
||||||
| NC score | 0.030961 (rank : 26) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q6A009, Q6PIW6, Q8BZ44 | Gene names | Znf294, Kiaa0714, zfp294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 294 (Zfp-294). | |||||
|
ZN294_HUMAN
|
||||||
| NC score | 0.030897 (rank : 27) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.029888 (rank : 28) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.029431 (rank : 29) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
NU214_HUMAN
|
||||||
| NC score | 0.027715 (rank : 30) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35658 | Gene names | NUP214, CAIN, CAN | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear pore complex protein Nup214 (Nucleoporin Nup214) (214 kDa nucleoporin) (CAN protein). | |||||
|
K0460_MOUSE
|
||||||
| NC score | 0.027113 (rank : 31) | θ value | 1.06291 (rank : 19) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6NXI6, Q3U3L8, Q6ZQA7 | Gene names | Kiaa0460 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0460. | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.026784 (rank : 32) | θ value | 0.813845 (rank : 16) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.026655 (rank : 33) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.026208 (rank : 34) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
CIP4_HUMAN
|
||||||
| NC score | 0.024726 (rank : 35) | θ value | 1.06291 (rank : 18) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15642, O15184, Q5TZN1 | Gene names | TRIP10, CIP4 | |||
|
Domain Architecture |

|
|||||
| Description | Cdc42-interacting protein 4 (Thyroid receptor-interacting protein 10) (TRIP-10). | |||||
|
RAPSN_MOUSE
|
||||||
| NC score | 0.024275 (rank : 36) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
RAPSN_HUMAN
|
||||||
| NC score | 0.023839 (rank : 37) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
CSRP1_HUMAN
|
||||||
| NC score | 0.022654 (rank : 38) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_MOUSE
|
||||||
| NC score | 0.022541 (rank : 39) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
PTER_HUMAN
|
||||||
| NC score | 0.022321 (rank : 40) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BW5, Q9BY46 | Gene names | PTER | |||
|
Domain Architecture |

|
|||||
| Description | Phosphotriesterase-related protein (Parathion hydrolase-related protein) (HPHRP). | |||||
|
GORS2_MOUSE
|
||||||
| NC score | 0.022195 (rank : 41) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99JX3, Q3U9D2, Q8CCD0, Q91W68 | Gene names | Gorasp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi reassembly-stacking protein 2 (GRS2) (Golgi reassembly-stacking protein of 55 kDa) (GRASP55). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.022178 (rank : 42) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.021233 (rank : 43) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.020601 (rank : 44) | θ value | 5.27518 (rank : 48) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
FOXM1_HUMAN
|
||||||
| NC score | 0.020022 (rank : 45) | θ value | 0.62314 (rank : 14) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q08050, O43258, O43259, O43260, Q4ZGG7, Q9BRL2 | Gene names | FOXM1, FKHL16, HFH11, MPP2, WIN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein M1 (Forkhead-related protein FKHL16) (Hepatocyte nuclear factor 3 forkhead homolog 11) (HNF-3/fork-head homolog 11) (HFH-11) (Winged helix factor from INS-1 cells) (M-phase phosphoprotein 2) (MPM-2 reactive phosphoprotein 2) (Transcription factor Trident). | |||||
|
CRSP7_HUMAN
|
||||||
| NC score | 0.019390 (rank : 46) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O95402 | Gene names | CRSP7, ARC70 | |||
|
Domain Architecture |

|
|||||
| Description | CRSP complex subunit 7 (Cofactor required for Sp1 transcriptional activation subunit 7) (Transcriptional coactivator CRSP70) (Activator- recruited cofactor 70 kDa component) (ARC70). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.019383 (rank : 47) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
ZN318_HUMAN
|
||||||
| NC score | 0.019072 (rank : 48) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5VUA4, O94796, Q4G0E4, Q8NEM6, Q9UNU8, Q9Y2W9 | Gene names | ZNF318 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 318 (Endocrine regulatory protein). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.018783 (rank : 49) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
CSRP2_MOUSE
|
||||||
| NC score | 0.018388 (rank : 50) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97314, Q9CZG5 | Gene names | Csrp2, Dlp1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Double LIM protein 1) (DLP-1). | |||||
|
CSRP2_HUMAN
|
||||||
| NC score | 0.018381 (rank : 51) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q16527, Q93030 | Gene names | CSRP2, LMO5, SMLIM | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 2 (Cysteine-rich protein 2) (CRP2) (Smooth muscle cell LIM protein) (SmLIM) (LIM-only protein 5). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.017430 (rank : 52) | θ value | 5.27518 (rank : 47) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
NUMB_HUMAN
|
||||||
| NC score | 0.016849 (rank : 53) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49757, Q6NUQ7, Q86SY1, Q8WW73, Q9UBG1, Q9UEQ4, Q9UKE8, Q9UKE9, Q9UKF0, Q9UQJ4 | Gene names | NUMB | |||
|
Domain Architecture |

|
|||||
| Description | Protein numb homolog (h-Numb) (Protein S171). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.015860 (rank : 54) | θ value | 5.27518 (rank : 46) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.015739 (rank : 55) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
RO52_MOUSE
|
||||||
| NC score | 0.015222 (rank : 56) | θ value | 1.06291 (rank : 20) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q62191 | Gene names | Trim21, Ro52, Ssa1 | |||
|
Domain Architecture |

|
|||||
| Description | 52 kDa Ro protein (Sjoegren syndrome type A antigen) (SS-A) (Ro(SS-A)) (52 kDa ribonucleoprotein autoantigen Ro/SS-A) (Tripartite motif- containing protein 21). | |||||
|
TBX10_HUMAN
|
||||||
| NC score | 0.014736 (rank : 57) | θ value | 0.47712 (rank : 13) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75333, Q86XS3 | Gene names | TBX10, TBX7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-box transcription factor TBX10 (T-box protein 10). | |||||
|
RAI1_HUMAN
|
||||||
| NC score | 0.012283 (rank : 58) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7Z5J4, Q8N3B4, Q8ND08, Q8WU64, Q96JK5, Q9H1C1, Q9H1C2, Q9UF69 | Gene names | RAI1, KIAA1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.011942 (rank : 59) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
EFNA4_MOUSE
|
||||||
| NC score | 0.011860 (rank : 60) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08542, O55218 | Gene names | Efna4, Epl4, Eplg4, Lerk4 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin-A4 precursor (EPH-related receptor tyrosine kinase ligand 4) (LERK-4). | |||||
|
CO4A_HUMAN
|
||||||
| NC score | 0.011002 (rank : 61) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P0C0L4, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q5JQM8, Q9NPK5, Q9UIP5 | Gene names | C4A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-A precursor (Acidic complement C4) [Contains: Complement C4 beta chain; Complement C4-A alpha chain; C4a anaphylatoxin; C4b-A; C4d-A; Complement C4 gamma chain]. | |||||
|
CO4B_HUMAN
|
||||||
| NC score | 0.010996 (rank : 62) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P0C0L5, P01028, P78445, Q13160, Q13906, Q14033, Q14835, Q9NPK5, Q9UIP5 | Gene names | C4B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C4-B precursor (Basic complement C4) [Contains: Complement C4 beta chain; Complement C4-B alpha chain; C4a anaphylatoxin; C4b-B; C4d-B; Complement C4 gamma chain]. | |||||
|
F100B_HUMAN
|
||||||
| NC score | 0.010931 (rank : 63) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IYN6 | Gene names | FAM100B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM100B. | |||||
|
PKHA6_MOUSE
|
||||||
| NC score | 0.010133 (rank : 64) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 591 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q7TQG1, Q8K0J5 | Gene names | Plekha6, Pepp3 | |||
|
Domain Architecture |

|
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
FOXL2_MOUSE
|
||||||
| NC score | 0.009854 (rank : 65) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88470, Q8K3X9 | Gene names | Foxl2, Pfrk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Forkhead box protein L2 (Pituitary forkhead factor) (P-Frk). | |||||
|
EFHD1_HUMAN
|
||||||
| NC score | 0.009712 (rank : 66) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BUP0, Q9BTF8, Q9H8I2, Q9HBQ0 | Gene names | EFHD1, SWS2 | |||
|
Domain Architecture |

|
|||||
| Description | EF-hand domain-containing protein 1 (Swiprosin-2). | |||||
|
ZCC2_HUMAN
|
||||||
| NC score | 0.009533 (rank : 67) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z2W4, Q8IW57, Q8TAJ3, Q96N79, Q9H8R9, Q9P0Y7 | Gene names | ZC3HAV1, ZC3HDC2 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger CCCH type antiviral protein 1 (Zinc finger CCCH domain- containing protein 2). | |||||
|
TTLL4_HUMAN
|
||||||
| NC score | 0.009490 (rank : 68) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14679, Q8WW29 | Gene names | TTLL4, KIAA0173 | |||
|
Domain Architecture |

|
|||||
| Description | Tubulin--tyrosine ligase-like protein 4. | |||||
|
GAB1_MOUSE
|
||||||
| NC score | 0.009463 (rank : 69) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QYY0, Q91VW7 | Gene names | Gab1 | |||
|
Domain Architecture |

|
|||||
| Description | GRB2-associated-binding protein 1 (GRB2-associated binder 1). | |||||
|
JHD2B_HUMAN
|
||||||
| NC score | 0.009287 (rank : 70) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 171 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q7LBC6, Q9BVH6, Q9BW93, Q9BZ52, Q9NYF4, Q9UPS0 | Gene names | JMJD1B, C5orf7, JHDM2B, KIAA1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B) (Nuclear protein 5qNCA). | |||||
|
C1TC_MOUSE
|
||||||
| NC score | 0.009271 (rank : 71) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q922D8 | Gene names | Mthfd1 | |||
|
Domain Architecture |

|
|||||
| Description | C-1-tetrahydrofolate synthase, cytoplasmic (C1-THF synthase) [Includes: Methylenetetrahydrofolate dehydrogenase (EC 1.5.1.5); Methenyltetrahydrofolate cyclohydrolase (EC 3.5.4.9); Formyltetrahydrofolate synthetase (EC 6.3.4.3)]. | |||||
|
KI2L4_HUMAN
|
||||||
| NC score | 0.008331 (rank : 72) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99706, O14621, O14622, O14623, O14624, O43534, P78400, P78401, Q99559, Q99560, Q99561, Q99562, Q9UQJ7 | Gene names | KIR2DL4, CD158D, KIR103AS | |||
|
Domain Architecture |

|
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL4 precursor (MHC class I NK cell receptor KIR103AS) (Killer cell inhibitory receptor 103AS) (KIR-103AS) (G9P) (CD158d antigen). | |||||
|
FOXL2_HUMAN
|
||||||
| NC score | 0.008270 (rank : 73) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58012, Q4ZGJ3 | Gene names | FOXL2 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein L2. | |||||
|
CAC1H_MOUSE
|
||||||
| NC score | 0.007717 (rank : 74) | θ value | 5.27518 (rank : 49) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 230 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O88427, Q80TJ2, Q9JKU5 | Gene names | Cacna1h, Kiaa1120 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1H (Voltage- gated calcium channel subunit alpha Cav3.2). | |||||
|
ARNT_MOUSE
|
||||||
| NC score | 0.007490 (rank : 75) | θ value | 5.27518 (rank : 45) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P53762, Q60661 | Gene names | Arnt | |||
|
Domain Architecture |

|
|||||
| Description | Aryl hydrocarbon receptor nuclear translocator (ARNT protein) (Dioxin receptor, nuclear translocator) (Hypoxia-inducible factor 1 beta) (HIF-1 beta). | |||||
|
EHMT1_HUMAN
|
||||||
| NC score | 0.006704 (rank : 76) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 417 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H9B1, Q8TCN7, Q96F53, Q96JF1, Q96KH4 | Gene names | EHMT1, EUHMTASE1, KIAA1876 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 5) (H3-K9-HMTase 5) (Euchromatic histone-lysine N-methyltransferase 1) (Eu-HMTase1) (G9a- like protein 1) (GLP1). | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.006454 (rank : 77) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
MAST4_HUMAN
|
||||||
| NC score | 0.005009 (rank : 78) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1804 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O15021, Q6ZN07, Q96LY3 | Gene names | MAST4, KIAA0303 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated serine/threonine-protein kinase 4 (EC 2.7.11.1). | |||||
|
ULK1_MOUSE
|
||||||
| NC score | 0.003349 (rank : 79) | θ value | 0.813845 (rank : 17) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1214 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O70405, Q6PGB2 | Gene names | Ulk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1) (Serine/threonine-protein kinase Unc51.1). | |||||
|
GLI1_MOUSE
|
||||||
| NC score | 0.002510 (rank : 80) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P47806, Q9QYK1 | Gene names | Gli1, Gli | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein GLI1 (Glioma-associated oncogene homolog). | |||||
|
M4K4_MOUSE
|
||||||
| NC score | 0.000552 (rank : 81) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 76 | Target Neighborhood Hits | 1560 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97820 | Gene names | Map4k4, Nik | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase kinase 4 (EC 2.7.11.1) (MAPK/ERK kinase kinase kinase 4) (MEK kinase kinase 4) (MEKKK 4) (HPK/GCK-like kinase HGK) (Nck-interacting kinase). | |||||