Please be patient as the page loads
|
DTX2_HUMAN
|
||||||
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |
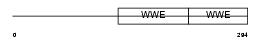
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DTX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
DTX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.985233 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |
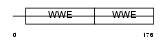
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
DTX1_HUMAN
|
||||||
| θ value | 6.60789e-144 (rank : 3) | NC score | 0.955685 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86Y01, O60630, Q9BS04 | Gene names | DTX1 | |||
|
Domain Architecture |
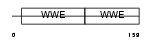
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (hDTX1). | |||||
|
DTX1_MOUSE
|
||||||
| θ value | 1.12714e-143 (rank : 4) | NC score | 0.947690 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |
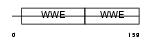
|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
DTX3L_HUMAN
|
||||||
| θ value | 8.89434e-40 (rank : 5) | NC score | 0.733967 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TDB6 | Gene names | DTX3L, BBAP | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2). | |||||
|
DTX3_HUMAN
|
||||||
| θ value | 1.57e-36 (rank : 6) | NC score | 0.699124 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N9I9, Q8NAU6, Q8NDS8 | Gene names | DTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3). | |||||
|
DTX3_MOUSE
|
||||||
| θ value | 4.568e-36 (rank : 7) | NC score | 0.695795 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80V91, Q9ER06 | Gene names | Dtx3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3) (mDTX3). | |||||
|
RNF32_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 8) | NC score | 0.164203 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
CIC_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.051900 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ENAH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.039509 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 11) | NC score | 0.043796 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
TRIPC_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.049647 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
TTC3_HUMAN
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.047970 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
MLL3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 14) | NC score | 0.039596 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 0.279714 (rank : 15) | NC score | 0.039846 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.040604 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
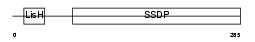
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.040604 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
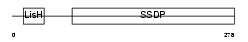
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
TULP4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.033788 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
ALMS1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.037390 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
ZFY28_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.035394 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
BCL9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.035216 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
HUWE1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.027936 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
HUWE1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 23) | NC score | 0.028474 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
RNF32_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.121262 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.031974 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.045874 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.045799 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
IER2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.030893 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
SMRA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.037982 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
SODM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.030017 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09671, Q64670, Q8VEM5 | Gene names | Sod2, Sod-2 | |||
|
Domain Architecture |
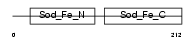
|
|||||
| Description | Superoxide dismutase [Mn], mitochondrial precursor (EC 1.15.1.1). | |||||
|
SRF_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.035409 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11831 | Gene names | SRF | |||
|
Domain Architecture |

|
|||||
| Description | Serum response factor (SRF). | |||||
|
ZDH14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.013908 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZN3, Q5JS07, Q5JS08, Q6PHS4, Q8IZN2, Q9H7F1 | Gene names | ZDHHC14 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.032132 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CT128_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.026247 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
DYR1B_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.001302 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z188 | Gene names | Dyrk1b | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1). | |||||
|
MKRN2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.029504 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
MLL3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.037605 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
PAX2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.016024 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02962, Q15105, Q15110, Q15837 | Gene names | PAX2 | |||
|
Domain Architecture |
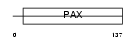
|
|||||
| Description | Paired box protein Pax-2. | |||||
|
PAX2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 39) | NC score | 0.016073 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32114 | Gene names | Pax2, Pax-2 | |||
|
Domain Architecture |
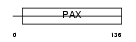
|
|||||
| Description | Paired box protein Pax-2. | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 2.36792 (rank : 40) | NC score | 0.035440 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | 2.36792 (rank : 41) | NC score | 0.025997 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TRAIP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 42) | NC score | 0.035424 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
BRCA1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.046539 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BRCA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.046390 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.032992 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
JADE2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.016209 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
KI2S1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 47) | NC score | 0.010754 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14954, O43471 | Gene names | KIR2DS1, CD158H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS1 precursor (MHC class I NK cell receptor Eb6 ActI) (CD158h antigen). | |||||
|
PELP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 48) | NC score | 0.047020 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 3.0926 (rank : 49) | NC score | 0.013822 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
LAMP3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.019334 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQV4, O94781, Q8NEC8 | Gene names | LAMP3, DCLAMP, TSC403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (Protein TSC403) (CD208 antigen). | |||||
|
MILK2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.017517 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
SMRC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.014202 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
SRF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.030794 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM73 | Gene names | Srf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum response factor (SRF). | |||||
|
BAZ1B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.021354 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
BAZ1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.022600 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
K0174_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.017853 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53990, Q9BQ81, Q9BWN2 | Gene names | KIAA0174 | |||
|
Domain Architecture |
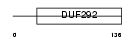
|
|||||
| Description | Protein KIAA0174 (Putative MAPK-activating protein PM28). | |||||
|
KLF5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | -0.000328 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||
|
MKRN2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.024303 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.017582 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
RM43_HUMAN
|
||||||
| θ value | 5.27518 (rank : 60) | NC score | 0.020904 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N983, Q7Z719, Q7Z7H6, Q86XN1, Q9BYC7 | Gene names | MRPL43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L43, mitochondrial precursor (L43mt) (MRP-L43) (Mitochondrial ribosomal protein bMRP36a). | |||||
|
SMRA3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 61) | NC score | 0.030694 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 62) | NC score | 0.030219 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.046333 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
HDAC5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.008326 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
K1543_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.026006 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
KI2L1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.008988 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
PTER_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.022047 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BW5, Q9BY46 | Gene names | PTER | |||
|
Domain Architecture |

|
|||||
| Description | Phosphotriesterase-related protein (Parathion hydrolase-related protein) (HPHRP). | |||||
|
TRI30_MOUSE
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.029115 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15533, Q99K55, Q99PQ7, Q99PQ8, Q99PQ9 | Gene names | Trim30, Rpt-1, Rpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 30 (Down regulatory protein of interleukin 2 receptor). | |||||
|
ZN294_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.029932 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
ZN294_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.029700 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6A009, Q6PIW6, Q8BZ44 | Gene names | Znf294, Kiaa0714, zfp294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 294 (Zfp-294). | |||||
|
41_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.003612 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
CBP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.015620 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
CL028_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.017484 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96LU7 | Gene names | C12orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf28 precursor. | |||||
|
CSRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.012847 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012770 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.022032 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
HDAC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.008412 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
LIRB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.006983 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
RAPSN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.022501 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |
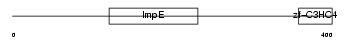
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
RAPSN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.023061 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |
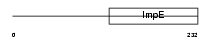
|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.010366 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
TOPRS_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.071262 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TULP4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.025674 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
WDFY3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.006143 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
RING1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.056610 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |
|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.055529 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
RN165_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.058498 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
TOPRS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.063768 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
DTX2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
DTX2_MOUSE
|
||||||
| NC score | 0.985233 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8R3P2, Q9CZV3, Q9ER07, Q9ER08 | Gene names | Dtx2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (mDTX2). | |||||
|
DTX1_HUMAN
|
||||||
| NC score | 0.955685 (rank : 3) | θ value | 6.60789e-144 (rank : 3) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q86Y01, O60630, Q9BS04 | Gene names | DTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (hDTX1). | |||||
|
DTX1_MOUSE
|
||||||
| NC score | 0.947690 (rank : 4) | θ value | 1.12714e-143 (rank : 4) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q61010, Q3TER5, Q8C2H2, Q9ER09 | Gene names | Dtx1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-1 (Deltex-1) (Deltex1) (mDTX1) (FXI-T1). | |||||
|
DTX3L_HUMAN
|
||||||
| NC score | 0.733967 (rank : 5) | θ value | 8.89434e-40 (rank : 5) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TDB6 | Gene names | DTX3L, BBAP | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex 3-like protein (B-lymphoma- and BAL-associated protein) (Rhysin-2) (Rhysin2). | |||||
|
DTX3_HUMAN
|
||||||
| NC score | 0.699124 (rank : 6) | θ value | 1.57e-36 (rank : 6) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 198 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8N9I9, Q8NAU6, Q8NDS8 | Gene names | DTX3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3). | |||||
|
DTX3_MOUSE
|
||||||
| NC score | 0.695795 (rank : 7) | θ value | 4.568e-36 (rank : 7) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80V91, Q9ER06 | Gene names | Dtx3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-3 (Deltex-3) (Deltex3) (mDTX3). | |||||
|
RNF32_HUMAN
|
||||||
| NC score | 0.164203 (rank : 8) | θ value | 0.00390308 (rank : 8) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9H0A6, Q6FIB3, Q6X7T4, Q8N6V8, Q8TDG0, Q96BM5, Q9Y6U1 | Gene names | RNF32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32. | |||||
|
RNF32_MOUSE
|
||||||
| NC score | 0.121262 (rank : 9) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9JIT1, Q9D9X0, Q9DAJ3 | Gene names | Rnf32, Lmbr2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 32 (Limb region protein 2). | |||||
|
TOPRS_MOUSE
|
||||||
| NC score | 0.071262 (rank : 10) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80Z37, Q3U5B5, Q3UPZ1, Q3USN3, Q3UZ55, Q8BXP2, Q8CFF5, Q8CGC8, Q920L3 | Gene names | Topors | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
TOPRS_HUMAN
|
||||||
| NC score | 0.063768 (rank : 11) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9NS56, O43273, Q6P987, Q9NS55, Q9UNR9 | Gene names | TOPORS, LUN, TP53BPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein E3 ligase Topors (EC 6.3.2.-) (SUMO1-protein E3 ligase Topors) (Topoisomerase I-binding RING finger protein) (Topoisomerase I-binding arginine/serine-rich protein) (Tumor suppressor p53-binding protein 3) (p53-binding protein 3) (p53BP3). | |||||
|
RN165_HUMAN
|
||||||
| NC score | 0.058498 (rank : 12) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6ZSG1 | Gene names | RNF165 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 165. | |||||
|
RING1_HUMAN
|
||||||
| NC score | 0.056610 (rank : 13) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06587, Q5JP96, Q5SQW2, Q86V19 | Gene names | RING1, RNF1 | |||
|
Domain Architecture |

|
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1). | |||||
|
RING1_MOUSE
|
||||||
| NC score | 0.055529 (rank : 14) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O35730, Q3U242, Q3U333, Q4FK33, Q63ZX8, Q921Z8 | Gene names | Ring1, Ring1A, Rnf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb complex protein RING1 (RING finger protein 1) (Transcription repressor Ring1A). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.051900 (rank : 15) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
TRIPC_HUMAN
|
||||||
| NC score | 0.049647 (rank : 16) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q14669, Q15644 | Gene names | TRIP12, KIAA0045 | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid receptor-interacting protein 12 (TRIP12). | |||||
|
TTC3_HUMAN
|
||||||
| NC score | 0.047970 (rank : 17) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 664 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P53804, O60767, P78476, P78477 | Gene names | TTC3, TPRD | |||
|
Domain Architecture |

|
|||||
| Description | Tetratricopeptide repeat protein 3 (TPR repeat protein 3) (TPR repeat protein D). | |||||
|
PELP1_MOUSE
|
||||||
| NC score | 0.047020 (rank : 18) | θ value | 3.0926 (rank : 48) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9DBD5, Q5F2E2, Q6PEM0, Q91YM9 | Gene names | Pelp1, Mnar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-, glutamic acid- and leucine-rich protein 1 (Modulator of nongenomic activity of estrogen receptor). | |||||
|
BRCA1_HUMAN
|
||||||
| NC score | 0.046539 (rank : 19) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 381 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P38398 | Gene names | BRCA1, RNF53 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein (RING finger protein 53). | |||||
|
BRCA1_MOUSE
|
||||||
| NC score | 0.046390 (rank : 20) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P48754, Q60957, Q60983 | Gene names | Brca1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 1 susceptibility protein homolog. | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.046333 (rank : 21) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.045874 (rank : 22) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.045799 (rank : 23) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.043796 (rank : 24) | θ value | 0.0736092 (rank : 11) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.040604 (rank : 25) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.040604 (rank : 26) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.039846 (rank : 27) | θ value | 0.279714 (rank : 15) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
MLL3_HUMAN
|
||||||
| NC score | 0.039596 (rank : 28) | θ value | 0.279714 (rank : 14) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1644 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8NEZ4, Q8NC02, Q8NDF6, Q9H9P4, Q9NR13, Q9P222, Q9UDR7 | Gene names | MLL3, HALR, KIAA1506 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3) (Homologous to ALR protein). | |||||
|
ENAH_MOUSE
|
||||||
| NC score | 0.039509 (rank : 29) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q03173, P70430, P70431, P70432, P70433, Q5D053 | Gene names | Enah, Mena, Ndpp1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein enabled homolog (NPC-derived proline-rich protein 1) (NDPP-1). | |||||
|
SMRA3_HUMAN
|
||||||
| NC score | 0.037982 (rank : 30) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 265 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14527, Q14536, Q16051, Q7KYJ6, Q86YA5, Q92652, Q96KM9 | Gene names | SMARCA3, HIP116A, HLTF, SNF2L3, ZBU1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (DNA-binding protein/plasminogen activator inhibitor 1 regulator) (Helicase-like transcription factor) (HIP116). | |||||
|
MLL3_MOUSE
|
||||||
| NC score | 0.037605 (rank : 31) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8BRH4, Q5YLV9, Q8BK12, Q8C6M3, Q923H5, Q923H6 | Gene names | Mll3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog (EC 2.1.1.43) (Histone-lysine N-methyltransferase, H3 lysine-4 specific MLL3). | |||||
|
ALMS1_HUMAN
|
||||||
| NC score | 0.037390 (rank : 32) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TCU4, Q53S05, Q580Q8, Q86VP9, Q9Y4G4 | Gene names | ALMS1, KIAA0328 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alstrom syndrome protein 1. | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.035440 (rank : 33) | θ value | 2.36792 (rank : 40) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
TRAIP_MOUSE
|
||||||
| NC score | 0.035424 (rank : 34) | θ value | 2.36792 (rank : 42) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VIG6, O08854, Q922M8, Q9CPP4 | Gene names | Traip, Trip | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
SRF_HUMAN
|
||||||
| NC score | 0.035409 (rank : 35) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11831 | Gene names | SRF | |||
|
Domain Architecture |

|
|||||
| Description | Serum response factor (SRF). | |||||
|
ZFY28_HUMAN
|
||||||
| NC score | 0.035394 (rank : 36) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HCC9 | Gene names | ZFYVE28, KIAA1643 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger FYVE domain-containing protein 28. | |||||
|
BCL9_HUMAN
|
||||||
| NC score | 0.035216 (rank : 37) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1008 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O00512 | Gene names | BCL9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | B-cell lymphoma 9 protein (Bcl-9) (Legless homolog). | |||||
|
TULP4_HUMAN
|
||||||
| NC score | 0.033788 (rank : 38) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NRJ4, Q9HD22, Q9P2F0 | Gene names | TULP4, KIAA1397, TUSP | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.032992 (rank : 39) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.032132 (rank : 40) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.031974 (rank : 41) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
IER2_HUMAN
|
||||||
| NC score | 0.030893 (rank : 42) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BTL4, Q03827, Q2TAZ2 | Gene names | IER2, ETR101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immediate early response gene 2 protein (Protein ETR101). | |||||
|
SRF_MOUSE
|
||||||
| NC score | 0.030794 (rank : 43) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JM73 | Gene names | Srf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum response factor (SRF). | |||||
|
SMRA3_MOUSE
|
||||||
| NC score | 0.030694 (rank : 44) | θ value | 5.27518 (rank : 61) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6PCN7, O35596, O35597, Q80VT6 | Gene names | Smarca3, Snf2l3, Zbu1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 3) (TNF-response element-binding protein) (P113). | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.030219 (rank : 45) | θ value | 5.27518 (rank : 62) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
SODM_MOUSE
|
||||||
| NC score | 0.030017 (rank : 46) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 3 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09671, Q64670, Q8VEM5 | Gene names | Sod2, Sod-2 | |||
|
Domain Architecture |

|
|||||
| Description | Superoxide dismutase [Mn], mitochondrial precursor (EC 1.15.1.1). | |||||
|
ZN294_HUMAN
|
||||||
| NC score | 0.029932 (rank : 47) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
ZN294_MOUSE
|
||||||
| NC score | 0.029700 (rank : 48) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6A009, Q6PIW6, Q8BZ44 | Gene names | Znf294, Kiaa0714, zfp294 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 294 (Zfp-294). | |||||
|
MKRN2_MOUSE
|
||||||
| NC score | 0.029504 (rank : 49) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9ERV1, Q9D0L9 | Gene names | Mkrn2 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2. | |||||
|
TRI30_MOUSE
|
||||||
| NC score | 0.029115 (rank : 50) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 434 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P15533, Q99K55, Q99PQ7, Q99PQ8, Q99PQ9 | Gene names | Trim30, Rpt-1, Rpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 30 (Down regulatory protein of interleukin 2 receptor). | |||||
|
HUWE1_MOUSE
|
||||||
| NC score | 0.028474 (rank : 51) | θ value | 0.813845 (rank : 23) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q7TMY8, Q4G2Z1, Q5BMM7, Q6NS61, Q8BNJ7, Q8CFH2, Q8VD14, Q921M5, Q9R0P2 | Gene names | Huwe1, Kiaa0312, Ureb1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (E3Histone). | |||||
|
HUWE1_HUMAN
|
||||||
| NC score | 0.027936 (rank : 52) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z6Z7, O15029, Q4G2Z2, Q5H961, Q6P4D0, Q8NG67, Q9BUI0, Q9HCJ4, Q9NSL6, Q9P0A9 | Gene names | HUWE1, KIAA0312, KIAA1578, UREB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HECT, UBA and WWE domain-containing protein 1 (EC 6.3.2.-) (E3 ubiquitin protein ligase URE-B1) (Mcl-1 ubiquitin ligase E3) (Mule) (ARF-binding protein 1) (ARF-BP1). | |||||
|
CT128_HUMAN
|
||||||
| NC score | 0.026247 (rank : 53) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BQN1, Q5JWN6, Q8N276 | Gene names | C20orf128 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf128. | |||||
|
K1543_HUMAN
|
||||||
| NC score | 0.026006 (rank : 54) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9P1Y5, Q8NDF1 | Gene names | KIAA1543 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1543. | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.025997 (rank : 55) | θ value | 2.36792 (rank : 41) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
TULP4_MOUSE
|
||||||
| NC score | 0.025674 (rank : 56) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9JIL5, Q8CA75, Q922C2 | Gene names | Tulp4, Tusp | |||
|
Domain Architecture |

|
|||||
| Description | Tubby-like protein 4 (Tubby superfamily protein). | |||||
|
MKRN2_HUMAN
|
||||||
| NC score | 0.024303 (rank : 57) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H000, Q8N391, Q96BD4, Q9BUY2, Q9NRY1 | Gene names | MKRN2, RNF62 | |||
|
Domain Architecture |

|
|||||
| Description | Makorin-2 (RING finger protein 62). | |||||
|
RAPSN_MOUSE
|
||||||
| NC score | 0.023061 (rank : 58) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12672 | Gene names | Rapsn | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
BAZ1B_MOUSE
|
||||||
| NC score | 0.022600 (rank : 59) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Z277, Q9CU68 | Gene names | Baz1b, Wbscr9 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein homolog) (WBRS9). | |||||
|
RAPSN_HUMAN
|
||||||
| NC score | 0.022501 (rank : 60) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13702, Q9BTD9 | Gene names | RAPSN | |||
|
Domain Architecture |

|
|||||
| Description | 43 kDa receptor-associated protein of the synapse (RAPsyn) (Acetylcholine receptor-associated 43 kDa protein) (43 kDa postsynaptic protein). | |||||
|
PTER_HUMAN
|
||||||
| NC score | 0.022047 (rank : 61) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96BW5, Q9BY46 | Gene names | PTER | |||
|
Domain Architecture |

|
|||||
| Description | Phosphotriesterase-related protein (Parathion hydrolase-related protein) (HPHRP). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.022032 (rank : 62) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
BAZ1B_HUMAN
|
||||||
| NC score | 0.021354 (rank : 63) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 701 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9UIG0, O95039, O95247, O95277 | Gene names | BAZ1B, WBSC10, WBSCR9, WSTF | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain adjacent to zinc finger domain protein 1B (Williams-Beuren syndrome chromosome region 9 protein) (Williams syndrome transcription factor) (hWALP2). | |||||
|
RM43_HUMAN
|
||||||
| NC score | 0.020904 (rank : 64) | θ value | 5.27518 (rank : 60) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N983, Q7Z719, Q7Z7H6, Q86XN1, Q9BYC7 | Gene names | MRPL43 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 39S ribosomal protein L43, mitochondrial precursor (L43mt) (MRP-L43) (Mitochondrial ribosomal protein bMRP36a). | |||||
|
LAMP3_HUMAN
|
||||||
| NC score | 0.019334 (rank : 65) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQV4, O94781, Q8NEC8 | Gene names | LAMP3, DCLAMP, TSC403 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Lysosome-associated membrane glycoprotein 3 precursor (LAMP-3) (Lysosomal-associated membrane protein 3) (DC-lysosome-associated membrane glycoprotein) (DC LAMP) (Protein TSC403) (CD208 antigen). | |||||
|
K0174_HUMAN
|
||||||
| NC score | 0.017853 (rank : 66) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53990, Q9BQ81, Q9BWN2 | Gene names | KIAA0174 | |||
|
Domain Architecture |

|
|||||
| Description | Protein KIAA0174 (Putative MAPK-activating protein PM28). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.017582 (rank : 67) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
MILK2_HUMAN
|
||||||
| NC score | 0.017517 (rank : 68) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IY33, Q7RTP4, Q7Z655, Q8TEQ4, Q9H5F9 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | MICAL-like protein 2. | |||||
|
CL028_HUMAN
|
||||||
| NC score | 0.017484 (rank : 69) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96LU7 | Gene names | C12orf28 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C12orf28 precursor. | |||||
|
JADE2_MOUSE
|
||||||
| NC score | 0.016209 (rank : 70) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6ZQF7, Q3UHD5, Q6IE83 | Gene names | Phf15, Jade2, Kiaa0239 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein Jade-2 (PHD finger protein 15). | |||||
|
PAX2_MOUSE
|
||||||
| NC score | 0.016073 (rank : 71) | θ value | 2.36792 (rank : 39) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P32114 | Gene names | Pax2, Pax-2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-2. | |||||
|
PAX2_HUMAN
|
||||||
| NC score | 0.016024 (rank : 72) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q02962, Q15105, Q15110, Q15837 | Gene names | PAX2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-2. | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.015620 (rank : 73) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
SMRC1_MOUSE
|
||||||
| NC score | 0.014202 (rank : 74) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P97496, Q7TS80, Q7TT29 | Gene names | Smarcc1, Baf155, Srg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SWI/SNF complex 155 kDa subunit) (BRG1-associated factor 155) (SWI3-related protein). | |||||
|
ZDH14_HUMAN
|
||||||
| NC score | 0.013908 (rank : 75) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8IZN3, Q5JS07, Q5JS08, Q6PHS4, Q8IZN2, Q9H7F1 | Gene names | ZDHHC14 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC14 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 14) (DHHC-14) (NEW1 domain-containing protein) (NEW1CP). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.013822 (rank : 76) | θ value | 3.0926 (rank : 49) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
CSRP1_HUMAN
|
||||||
| NC score | 0.012847 (rank : 77) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P21291 | Gene names | CSRP1, CSRP, CYRP | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
CSRP1_MOUSE
|
||||||
| NC score | 0.012770 (rank : 78) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97315 | Gene names | Csrp1, Crp1, Csrp | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine and glycine-rich protein 1 (Cysteine-rich protein 1) (CRP1) (CRP). | |||||
|
KI2S1_HUMAN
|
||||||
| NC score | 0.010754 (rank : 79) | θ value | 3.0926 (rank : 47) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14954, O43471 | Gene names | KIR2DS1, CD158H | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DS1 precursor (MHC class I NK cell receptor Eb6 ActI) (CD158h antigen). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.010366 (rank : 80) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
KI2L1_HUMAN
|
||||||
| NC score | 0.008988 (rank : 81) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43626, O43470 | Gene names | KIR2DL1, CD158A, NKAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Killer cell immunoglobulin-like receptor 2DL1 precursor (MHC class I NK cell receptor) (Natural killer-associated transcript 1) (NKAT-1) (p58 natural killer cell receptor clones CL-42/47.11) (p58 NK receptor) (p58.1 MHC class-I-specific NK receptor) (CD158a antigen). | |||||
|
HDAC5_MOUSE
|
||||||
| NC score | 0.008412 (rank : 82) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2V6, Q9JL73 | Gene names | Hdac5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Histone deacetylase mHDA1). | |||||
|
HDAC5_HUMAN
|
||||||
| NC score | 0.008326 (rank : 83) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQL6, O60340, O60528, Q96DY4 | Gene names | HDAC5, KIAA0600 | |||
|
Domain Architecture |

|
|||||
| Description | Histone deacetylase 5 (HD5) (Antigen NY-CO-9). | |||||
|
LIRB3_HUMAN
|
||||||
| NC score | 0.006983 (rank : 84) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75022, O15471, Q86U49 | Gene names | LILRB3, ILT5, LIR3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte immunoglobulin-like receptor subfamily B member 3 precursor (Leukocyte immunoglobulin-like receptor 3) (LIR-3) (Immunoglobulin- like transcript 5) (ILT-5) (Monocyte inhibitory receptor HL9) (CD85a antigen). | |||||
|
WDFY3_MOUSE
|
||||||
| NC score | 0.006143 (rank : 85) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6VNB8, Q8C8H7 | Gene names | Wdfy3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WD repeat and FYVE domain-containing protein 3 (Beach domain, WD repeat and FYVE domain-containing protein 1) (BWF1). | |||||
|
41_HUMAN
|
||||||
| NC score | 0.003612 (rank : 86) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11171, P11176, Q14245, Q5TB35, Q5VXN8, Q8IXV9, Q9Y578, Q9Y579 | Gene names | EPB41, E41P | |||
|
Domain Architecture |

|
|||||
| Description | Protein 4.1 (Band 4.1) (P4.1) (EPB4.1) (4.1R). | |||||
|
DYR1B_MOUSE
|
||||||
| NC score | 0.001302 (rank : 87) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 799 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z188 | Gene names | Dyrk1b | |||
|
Domain Architecture |

|
|||||
| Description | Dual specificity tyrosine-phosphorylation-regulated kinase 1B (EC 2.7.12.1). | |||||
|
KLF5_HUMAN
|
||||||
| NC score | -0.000328 (rank : 88) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 84 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q13887, Q9UHP8 | Gene names | KLF5, BTEB2, CKLF, IKLF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Krueppel-like factor 5 (Intestinal-enriched krueppel-like factor) (Colon krueppel-like factor) (Transcription factor BTEB2) (Basic transcription element-binding protein 2) (BTE-binding protein 2) (GC box-binding protein 2). | |||||