Please be patient as the page loads
|
DAZP2_MOUSE
|
||||||
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DAZP2_MOUSE
|
||||||
| θ value | 3.4653e-68 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | 1.00825e-67 (rank : 2) | NC score | 0.999178 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 3) | NC score | 0.117080 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
RPB1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.111239 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.111132 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
CDX1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.042073 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18111, Q8VCF7 | Gene names | Cdx1, Cdx-1 | |||
|
Domain Architecture |
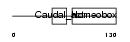
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.105752 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
PLS4_HUMAN
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.109062 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.104422 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
B4GT1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.053781 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
US6NL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.065766 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
CX045_HUMAN
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.120756 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5U8, Q5JXY9 | Gene names | CXorf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CXorf45. | |||||
|
CXA3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.028703 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64448 | Gene names | Gja3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
SFR15_HUMAN
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.064666 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
DTX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.045799 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |
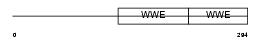
|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
HXA4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.024925 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |
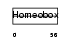
|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
LOXL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.032986 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.010760 (rank : 37) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
B3A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 19) | NC score | 0.015068 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
CDX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.028881 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |
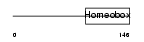
|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
YLPM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.039290 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
BAT2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.028681 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.018111 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
PLS4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.080201 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
T22D2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.039642 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.037371 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
RAI1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.027262 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.005952 (rank : 39) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
BAT3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 29) | NC score | 0.022972 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
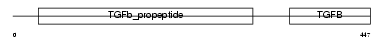
BMP5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.009707 (rank : 38) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49003 | Gene names | Bmp5, Bmp-5 | |||
|
Domain Architecture |
|
|||||
| Description | Bone morphogenetic protein 5 precursor (BMP-5). | |||||
|
ERBB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.004055 (rank : 40) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
GGN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.019818 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
HXB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.018388 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P14653 | Gene names | HOXB1, HOX2I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B1 (Hox-2I). | |||||
|
RTP3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.034382 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
ZN341_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.001522 (rank : 41) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYN7, Q5JXM8, Q96ST5 | Gene names | ZNF341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 341. | |||||
|
PLS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.053320 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.053614 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.053833 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
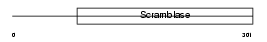
PLS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.053956 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |
|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
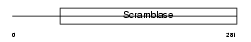
PLS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.061237 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |
|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
RPC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.062659 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 3.4653e-68 (rank : 1) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.999178 (rank : 2) | θ value | 1.00825e-67 (rank : 2) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
CX045_HUMAN
|
||||||
| NC score | 0.120756 (rank : 3) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H5U8, Q5JXY9 | Gene names | CXorf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CXorf45. | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.117080 (rank : 4) | θ value | 0.0736092 (rank : 3) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
RPB1_HUMAN
|
||||||
| NC score | 0.111239 (rank : 5) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P24928 | Gene names | POLR2A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
RPB1_MOUSE
|
||||||
| NC score | 0.111132 (rank : 6) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P08775 | Gene names | Polr2a, Rpii215, Rpo2-1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase II largest subunit (EC 2.7.7.6) (RPB1). | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.109062 (rank : 7) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.105752 (rank : 8) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.104422 (rank : 9) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
PLS4_MOUSE
|
||||||
| NC score | 0.080201 (rank : 10) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
US6NL_MOUSE
|
||||||
| NC score | 0.065766 (rank : 11) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q80XC3, Q6ZQK8 | Gene names | Usp6nl, Kiaa0019 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | USP6 N-terminal-like protein. | |||||
|
SFR15_HUMAN
|
||||||
| NC score | 0.064666 (rank : 12) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 368 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95104, Q6P1M5, Q8N3I8, Q9UFM1, Q9ULP8 | Gene names | SFRS15, KIAA1172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 15 (CTD-binding SR-like protein RA4). | |||||
|
RPC1_HUMAN
|
||||||
| NC score | 0.062659 (rank : 13) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O14802, Q8TCW5 | Gene names | POLR3A | |||
|
Domain Architecture |

|
|||||
| Description | DNA-directed RNA polymerase III largest subunit (EC 2.7.7.6) (RPC155) (RPC1). | |||||
|
PLS3_HUMAN
|
||||||
| NC score | 0.061237 (rank : 14) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS2_MOUSE
|
||||||
| NC score | 0.053956 (rank : 15) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS2_HUMAN
|
||||||
| NC score | 0.053833 (rank : 16) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
B4GT1_HUMAN
|
||||||
| NC score | 0.053781 (rank : 17) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P15291, Q12909, Q12910, Q12911, Q14456, Q14509, Q14523 | Gene names | B4GALT1, GGTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 1 (EC 2.4.1.-) (Beta-1,4-GalTase 1) (Beta4Gal-T1) (b4Gal-T1) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 1) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 1) [Includes: Lactose synthase A protein (EC 2.4.1.22); N-acetyllactosamine synthase (EC 2.4.1.90) (Nal synthetase); Beta-N-acetylglucosaminylglycopeptide beta-1,4- galactosyltransferase (EC 2.4.1.38); Beta-N-acetylglucosaminyl- glycolipid beta-1,4-galactosyltransferase (EC 2.4.1.-)]. | |||||
|
PLS1_MOUSE
|
||||||
| NC score | 0.053614 (rank : 18) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS1_HUMAN
|
||||||
| NC score | 0.053320 (rank : 19) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
DTX2_HUMAN
|
||||||
| NC score | 0.045799 (rank : 20) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UW9, Q96H69, Q9H890, Q9P200 | Gene names | DTX2, KIAA1528 | |||
|
Domain Architecture |

|
|||||
| Description | Protein deltex-2 (Deltex-2) (Deltex2) (hDTX2). | |||||
|
CDX1_MOUSE
|
||||||
| NC score | 0.042073 (rank : 21) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P18111, Q8VCF7 | Gene names | Cdx1, Cdx-1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
T22D2_HUMAN
|
||||||
| NC score | 0.039642 (rank : 22) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O75157, Q6PI50, Q9H2Z6, Q9H2Z7, Q9H2Z8 | Gene names | TSC22D2, KIAA0669, TILZ4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TSC22 domain family protein 2 (TSC22-related-inducible leucine zipper protein 4). | |||||
|
YLPM1_MOUSE
|
||||||
| NC score | 0.039290 (rank : 23) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 505 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9R0I7, Q7TMM4 | Gene names | Ylpm1, Zap, Zap3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | YLP motif-containing protein 1 (Nuclear protein ZAP3). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.037371 (rank : 24) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
RTP3_MOUSE
|
||||||
| NC score | 0.034382 (rank : 25) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5QGU6, Q8VDC2 | Gene names | Rtp3, Tmem7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor-transporting protein 3 (Transmembrane protein 7). | |||||
|
LOXL1_HUMAN
|
||||||
| NC score | 0.032986 (rank : 26) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q08397, Q96BW7 | Gene names | LOXL1, LOXL | |||
|
Domain Architecture |

|
|||||
| Description | Lysyl oxidase homolog 1 precursor (EC 1.4.3.-) (Lysyl oxidase-like protein 1) (LOL). | |||||
|
CDX1_HUMAN
|
||||||
| NC score | 0.028881 (rank : 27) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P47902, Q4VAU4, Q9NYK8 | Gene names | CDX1 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein CDX-1 (Caudal-type homeobox protein 1). | |||||
|
CXA3_MOUSE
|
||||||
| NC score | 0.028703 (rank : 28) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64448 | Gene names | Gja3 | |||
|
Domain Architecture |

|
|||||
| Description | Gap junction alpha-3 protein (Connexin-46) (Cx46). | |||||
|
BAT2_HUMAN
|
||||||
| NC score | 0.028681 (rank : 29) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P48634, O95875, Q5SQ29, Q5SQ30, Q5ST84, Q5STX6, Q5STX7, Q68DW9, Q6P9P7, Q6PIN1, Q96QC6 | Gene names | BAT2, G2 | |||
|
Domain Architecture |

|
|||||
| Description | Large proline-rich protein BAT2 (HLA-B-associated transcript 2). | |||||
|
RAI1_MOUSE
|
||||||
| NC score | 0.027262 (rank : 30) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61818, Q6ZPH7, Q8CEV1 | Gene names | Rai1, Kiaa1820 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoic acid-induced protein 1. | |||||
|
HXA4_MOUSE
|
||||||
| NC score | 0.024925 (rank : 31) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P06798, Q61684, Q64388, Q8BPE6 | Gene names | Hoxa4, Hox-1.4, Hoxa-4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A4 (Hox-1.4) (Homeobox protein MH-3). | |||||
|
BAT3_MOUSE
|
||||||
| NC score | 0.022972 (rank : 32) | θ value | 8.99809 (rank : 29) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Z1R2, Q8SNA3 | Gene names | Bat3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Large proline-rich protein BAT3 (HLA-B-associated transcript 3). | |||||
|
GGN_HUMAN
|
||||||
| NC score | 0.019818 (rank : 33) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86UU5, Q7RTU6, Q86UU4, Q8NAA1 | Gene names | GGN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
HXB1_HUMAN
|
||||||
| NC score | 0.018388 (rank : 34) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P14653 | Gene names | HOXB1, HOX2I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-B1 (Hox-2I). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.018111 (rank : 35) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
B3A3_HUMAN
|
||||||
| NC score | 0.015068 (rank : 36) | θ value | 4.03905 (rank : 19) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.010760 (rank : 37) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
BMP5_MOUSE
|
||||||
| NC score | 0.009707 (rank : 38) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49003 | Gene names | Bmp5, Bmp-5 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 5 precursor (BMP-5). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.005952 (rank : 39) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ERBB2_HUMAN
|
||||||
| NC score | 0.004055 (rank : 40) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P04626, Q14256, Q6LDV1, Q9UMK4 | Gene names | ERBB2, HER2, NEU, NGL | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-2 precursor (EC 2.7.10.1) (p185erbB2) (C-erbB-2) (NEU proto-oncogene) (Tyrosine kinase-type cell surface receptor HER2) (MLN 19). | |||||
|
ZN341_HUMAN
|
||||||
| NC score | 0.001522 (rank : 41) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 35 | Target Neighborhood Hits | 803 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYN7, Q5JXM8, Q96ST5 | Gene names | ZNF341 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 341. | |||||