Please be patient as the page loads
|
PLS3_HUMAN
|
||||||
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |
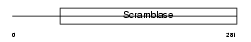
|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLS3_HUMAN
|
||||||
| θ value | 6.38546e-147 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| θ value | 1.86219e-138 (rank : 2) | NC score | 0.992401 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS1_MOUSE
|
||||||
| θ value | 4.22625e-74 (rank : 3) | NC score | 0.963116 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS1_HUMAN
|
||||||
| θ value | 2.73936e-73 (rank : 4) | NC score | 0.973613 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS2_MOUSE
|
||||||
| θ value | 6.98233e-69 (rank : 5) | NC score | 0.971388 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |
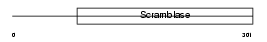
|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS2_HUMAN
|
||||||
| θ value | 2.03155e-68 (rank : 6) | NC score | 0.978945 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS4_HUMAN
|
||||||
| θ value | 1.51363e-47 (rank : 7) | NC score | 0.941087 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS4_MOUSE
|
||||||
| θ value | 1.85029e-45 (rank : 8) | NC score | 0.936982 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
COKA1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 9) | NC score | 0.026673 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
SPRR3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.066482 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.052328 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 12) | NC score | 0.013595 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CTGE4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.038208 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
SON_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.035637 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
CTGE6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.022792 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
SPR1A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.033396 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
NUMBL_HUMAN
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.023023 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.016765 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
FOXN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.017727 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
EGR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.002541 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||
|
FOXJ2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.025806 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
K0310_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.028073 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
MUC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.006734 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
TCF7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.010719 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
TPO_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.026130 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.011164 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
IASPP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.011479 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
OGFR_HUMAN
|
||||||
| θ value | 8.99809 (rank : 28) | NC score | 0.016857 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DAZP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.061322 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.061237 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
PLS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.38546e-147 (rank : 1) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NRY6, Q8NBW6, Q96F13 | Gene names | PLSCR3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS3_MOUSE
|
||||||
| NC score | 0.992401 (rank : 2) | θ value | 1.86219e-138 (rank : 2) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9JIZ9 | Gene names | Plscr3 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 3 (PL scramblase 3) (Ca(2+)-dependent phospholipid scramblase 3). | |||||
|
PLS2_HUMAN
|
||||||
| NC score | 0.978945 (rank : 3) | θ value | 2.03155e-68 (rank : 6) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NRY7 | Gene names | PLSCR2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_HUMAN
|
||||||
| NC score | 0.973613 (rank : 4) | θ value | 2.73936e-73 (rank : 4) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O15162 | Gene names | PLSCR1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Erythrocyte phospholipid scramblase) (MmTRA1b). | |||||
|
PLS2_MOUSE
|
||||||
| NC score | 0.971388 (rank : 5) | θ value | 6.98233e-69 (rank : 5) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCW2, O70233 | Gene names | Plscr2 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 2 (PL scramblase 2) (Ca(2+)-dependent phospholipid scramblase 2). | |||||
|
PLS1_MOUSE
|
||||||
| NC score | 0.963116 (rank : 6) | θ value | 4.22625e-74 (rank : 3) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9JJ00, O54730, O54731, Q9D1F8 | Gene names | Plscr1, Tra1b, Tras1 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 1 (PL scramblase 1) (Ca(2+)-dependent phospholipid scramblase 1) (Transplantability-associated protein 1) (TRA1) (NOR1). | |||||
|
PLS4_HUMAN
|
||||||
| NC score | 0.941087 (rank : 7) | θ value | 1.51363e-47 (rank : 7) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9NRQ2, Q7Z505 | Gene names | PLSCR4 | |||
|
Domain Architecture |

|
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4) (TRA1). | |||||
|
PLS4_MOUSE
|
||||||
| NC score | 0.936982 (rank : 8) | θ value | 1.85029e-45 (rank : 8) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P58196, Q3TMI2, Q8BH62 | Gene names | Plscr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phospholipid scramblase 4 (PL scramblase 4) (Ca(2+)-dependent phospholipid scramblase 4). | |||||
|
SPRR3_HUMAN
|
||||||
| NC score | 0.066482 (rank : 9) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 102 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UBC9, O75597, Q4ZGI7, Q5T525, Q8NET7, Q9UDG3 | Gene names | SPRR3, SPRC | |||
|
Domain Architecture |

|
|||||
| Description | Small proline-rich protein 3 (Cornifin beta) (Esophagin) (22 kDa pancornulin). | |||||
|
DAZP2_HUMAN
|
||||||
| NC score | 0.061322 (rank : 10) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15038 | Gene names | DAZAP2, KIAA0058 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2). | |||||
|
DAZP2_MOUSE
|
||||||
| NC score | 0.061237 (rank : 11) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DCP9, O88675, Q3UVI4 | Gene names | Dazap2, Prtb | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DAZ-associated protein 2 (Deleted in azoospermia-associated protein 2) (Proline-rich protein expressed in brain). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.052328 (rank : 12) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
CTGE4_HUMAN
|
||||||
| NC score | 0.038208 (rank : 13) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
SON_HUMAN
|
||||||
| NC score | 0.035637 (rank : 14) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P18583, O14487, O95981, Q14120, Q9H7B1, Q9P070, Q9P072, Q9UKP9, Q9UPY0 | Gene names | SON, C21orf50, DBP5, KIAA1019, NREBP | |||
|
Domain Architecture |

|
|||||
| Description | SON protein (SON3) (Negative regulatory element-binding protein) (NRE- binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1). | |||||
|
SPR1A_MOUSE
|
||||||
| NC score | 0.033396 (rank : 15) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q62266 | Gene names | Sprr1a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cornifin A (Small proline-rich protein 1A) (SPR1A) (SPR1 A). | |||||
|
K0310_HUMAN
|
||||||
| NC score | 0.028073 (rank : 16) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15027, Q5SXP0, Q5SXP1, Q8N347, Q96HP1 | Gene names | KIAA0310 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0310. | |||||
|
COKA1_HUMAN
|
||||||
| NC score | 0.026673 (rank : 17) | θ value | 0.21417 (rank : 9) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9P218, Q4VXQ4, Q8WUT2, Q96CY9, Q9BQU6, Q9BQU7 | Gene names | COL20A1, KIAA1510 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XX) chain precursor. | |||||
|
TPO_MOUSE
|
||||||
| NC score | 0.026130 (rank : 18) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P40226 | Gene names | Thpo | |||
|
Domain Architecture |

|
|||||
| Description | Thrombopoietin precursor (Megakaryocyte colony-stimulating factor) (Myeloproliferative leukemia virus oncogene ligand) (C-mpl ligand) (ML) (Megakaryocyte growth and development factor) (MGDF). | |||||
|
FOXJ2_HUMAN
|
||||||
| NC score | 0.025806 (rank : 19) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9P0K8, Q96PS9, Q9NSN5 | Gene names | FOXJ2, FHX | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J2 (Fork head homologous X). | |||||
|
NUMBL_HUMAN
|
||||||
| NC score | 0.023023 (rank : 20) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y6R0, Q7Z4J9 | Gene names | NUMBL | |||
|
Domain Architecture |

|
|||||
| Description | Numb-like protein (Numb-R). | |||||
|
CTGE6_HUMAN
|
||||||
| NC score | 0.022792 (rank : 21) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86UF2 | Gene names | CTAGE6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein cTAGE-6 (cTAGE family member 6). | |||||
|
FOXN1_HUMAN
|
||||||
| NC score | 0.017727 (rank : 22) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 377 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O15353, O15352 | Gene names | FOXN1, WHN | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein N1 (Transcription factor winged-helix nude). | |||||
|
OGFR_HUMAN
|
||||||
| NC score | 0.016857 (rank : 23) | θ value | 8.99809 (rank : 28) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1582 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9NZT2, O96029, Q4VXW5, Q96CM2, Q9BQW1, Q9H4H0, Q9H7J5, Q9NZT3, Q9NZT4 | Gene names | OGFR | |||
|
Domain Architecture |

|
|||||
| Description | Opioid growth factor receptor (OGFr) (Zeta-type opioid receptor) (7-60 protein). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.016765 (rank : 24) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.013595 (rank : 25) | θ value | 0.62314 (rank : 12) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
IASPP_MOUSE
|
||||||
| NC score | 0.011479 (rank : 26) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 1024 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5I1X5 | Gene names | Ppp1r13l, Nkip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RelA-associated inhibitor (Inhibitor of ASPP protein) (Protein iASPP) (PPP1R13B-like protein) (NFkB-interacting protein 1). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.011164 (rank : 27) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
TCF7_MOUSE
|
||||||
| NC score | 0.010719 (rank : 28) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q00417 | Gene names | Tcf7, Tcf-1, Tcf1 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor 7 (T-cell-specific transcription factor 1) (TCF- 1) (T-cell factor 1). | |||||
|
MUC1_HUMAN
|
||||||
| NC score | 0.006734 (rank : 29) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 975 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P15941, P13931, P15942, P17626, Q14128, Q14876, Q16437, Q16442, Q16615, Q9BXA4, Q9UE75, Q9UE76, Q9UQL1, Q9Y4J2 | Gene names | MUC1 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-1 precursor (MUC-1) (Polymorphic epithelial mucin) (PEM) (PEMT) (Episialin) (Tumor-associated mucin) (Carcinoma-associated mucin) (Tumor-associated epithelial membrane antigen) (EMA) (H23AG) (Peanut- reactive urinary mucin) (PUM) (Breast carcinoma-associated antigen DF3) (CD227 antigen). | |||||
|
EGR1_MOUSE
|
||||||
| NC score | 0.002541 (rank : 30) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 28 | Target Neighborhood Hits | 816 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08046, Q61777 | Gene names | Egr1, Egr-1, Krox-24 | |||
|
Domain Architecture |

|
|||||
| Description | Early growth response protein 1 (EGR-1) (Krox-24 protein) (Transcription factor Zif268) (Nerve growth factor-induced protein A) (NGFI-A). | |||||